En Face Endocardial Cushion Preparation for Planar Morphogenesis Analysis in Mouse Embryos
Summary
Classically, the endocardium of the mouse embryonic valve primordium has been analyzed using transversal, coronal, or sagittal sections. Our novel approach for en face, two-dimensional imaging of the endocardium at valvulogenic regions allows planar polarity and cell rearrangement analysis of the endocardium during valve development.
Abstract
The study of the cellular and molecular mechanisms underlying the development of the mammalian heart is essential to address human congenital heart disease. The development of the primitive cardiac valves involves the epithelial-to-mesenchymal transition (EMT) of endocardial cells from the atrioventricular canal (AVC) and outflow tract (OFT) regions of the heart in response to local inductive myocardial and endocardial signals. Once the cells delaminate and invade the extracellular matrix (cardiac jelly) located between the endocardium and the myocardium, the primitive endocardial cushions (EC) are formed. This process implies that the endocardium has to fill the gaps left by the delaminated cells and has to reorganize itself to converge (narrow) or extend (lengthen) along an axis. Current research has implicated the planar cell polarity (PCP) pathway in regulating the subcellular localization of the factors involved in this process. Classically, the initial phases of cardiac valve development have been studied in cross-sections of embryonic hearts or in ex vivo AVC or OFT explants cultured on collagen gels. These approaches allow the analysis of apico-basal polarity but do not allow the analysis of cell behavior within the plane of the epithelium or of the morphological changes of migrating cells. Here, we show an experimental approach that allows the visualization of the endocardium at valvulogenic regions as a planar field of cells. This experimental approach provides the opportunity to study PCP, planar topology, and intercellular communication within the endocardium of the OFT and AVC during valve development. Deciphering new cellular mechanisms involved in cardiac valve morphogenesis may contribute to understanding congenital heart disease associated with endocardial cushion defects.
Introduction
The heart is the first functional organ of a mammalian embryo. Around embryonic day (E) 7.5 in mice, bilateral precardiac mesoderm cells form the cardiac crescent in the ventral side1.The cardiac crescent contains two populations of precardiac cells that include progenitors of the myocardium and the endocardium2. Around E8.0, the cardiac precursors fuse in the midline, forming the primitive heart tube consisting of two epithelial tissues, the outer myocardium, and the inner endocardium, which is a specialized endothelium separated by an extracellular matrix named cardiac jelly. Later, at E8.5, the heart tube undergoes rightward looping. The looped heart has different anatomical regions with specific molecular signatures such as the outflow tract (OFT), the ventricles, and the atrio-ventricular canal (AVC)3. Although initially the heart tube expands at its inflow side through the addition of cells4, at E9.5, intensive cardiac proliferation results in ballooning of the chambers and establishment of the trabecular network5. Valve formation takes place in the AVC (future mitral and tricuspid valves) and in the OFT (future aortic and pulmonary valves).
The endocardium plays crucial roles in valve development. Endocardial cells undergo epithelial-mesenchymal transition (EMT) in the AVC and OFT to form the endocardial cushions, a structure that appears at the onset of valve development. Different signaling pathways activate this process; at E9.5 in mice, NOTCH activated in the endocardium in response to myocardial-derived BMP2 promotes invasive EMT of endocardial cells in the AVC and OFT regions through activation of TGFβ2 and SNAIL (SNAI1), which directly represses the expression of vascular endothelial cadherin (VE-cadherin), a transmembrane component of adherens junctions (AJs)6,7,8. In the OFT, activation of the endocardium to initiate EMT is mediated by FGF8 and BMP4, whose expression is activated by NOTCH9,10,11,12.
Progression of EMT involves cellular dynamics as cells change shape, break and remake junctions with their neighbors, delaminate, and begin to migrate13. These changes include AJ remodel and gradual disassembling14,15, planar cell polarity (PCP) signaling, the loss of apico-basal polarity (ABP), apical constriction, and cytoskeletal organization16,17. ABP refers to the distribution of proteins along the anterior-posterior axis of a cell. In the developing heart, ABP regulation in cardiomyocytes is required for ventricular development18. PCP refers to a polarized distribution of proteins within cells across the plane of a tissue and regulates cellular distribution; epithelia with a stable geometry are made up of hexagon-shaped cells, where only three cells converge at the vertices19,20,21,22. Different cellular processes, such as cell division, neighbor exchange, or delamination occurring during epithelial morphogenesis, produce an increase in the number of cells that converge on a vertex and the number of neighboring cells that a given cell has22. These cellular behaviors related to PCP can be regulated by different signaling pathways, actin dynamics, or intracellular trafficking23.
The data generated studying valve development in mice have been obtained from transversal, coronal, or sagittal sections of E8.5 and E9.5 embryonic hearts, where the endocardium is shown as a line of cells instead of as a field of cells-the endocardium covers the entire inner surface of the heart tube24. Embryonic sections do not allow the analysis of PCP in the endocardium of mouse embryos. Our novel experimental method allows the analysis of endocardial cell distribution, AJ anisotropy, and single-cell shape analysis, as shown in the representative results. This type of data is required for PCP analysis, together with the description of other molecules related to PCP, not shown in this report. Whole-mount immunofluorescence, specific sample preparation and the use of genetically modified mice enable planar polarity analysis in the endocardium at the onset of valve development in mice.
Protocol
Animal studies were approved by the Centro Nacional de Investigaciones Cardiovasculares (CNIC) Animal Experimentation Ethics Committee and by the Community of Madrid (ref. PROEX 155.7/20). All animal procedures conformed to EU Directive 2010/63EU and Recommendation 2007/526/EC regarding the protection of animals used for experimental and other scientific purposes, enacted in Spanish law under Real Decreto 1201/2005.
1. Obtention of AVC and/or OFTs (adapted from Xiao et al.25)
- Set up crosses in the afternoon between female mTmG and male VE-Cadherin-Cre-ERT/+.
- The next morning, check for a vaginal plug. If there is a vaginal plug, count it as half a day (E0.5). Depending on the experiment of interest, count 8 days or 9 days.
- 24 h before dissection, gavage the pregnant female orally with 50 µL of 5 mg/mL 4-OH-Tamoxifen diluted in corn oil.
NOTE: This dose will induce a limited amount of CRE-mediated recombination revealed by the presence of GFP-positive clones. The appropriate dose will have to be determined with each new batch of 4-OH-Tamoxifen. - Sacrifice the pregnant female at E8.5 or E9.5 by cervical dislocation.
- Open the abdomen of the pregnant mice by using scissors and remove the uterus containing the embryos by using scissors and fine forceps.
- Place the uterus in a Petri dish containing ice-cold 1x phosphate-buffered saline (PBS) with 0.1% Tween-20 (PBT).
- Remove the individual deciduae from the uterus under a dissecting microscope and using fine forceps.
- Using fine tweezers, make an incision in the white part of the decidua, which is where the embryo is, remove it gently, avoiding pulling, and transfer the embryos to a new Petri dish with fresh PBT using a P1000 with the tip cut off, leaving a diameter large enough for an embryo to pass through without breaking.
- For genotyping, take a little piece of the yolk sac (0.5 mm2) or a piece of the tail (from the posterior end, counting 5 to 10 somites toward the head) and digest it in 100 µL of appropriate buffer.
- Under the fume hood, transfer the embryos to ice-cold 4% paraformaldehyde (4% PFA) in sterile-filtered PBS in a microcentrifuge (2 mL) tube at 4 °C. Fix the embryos from 2 h to overnight (O/N) at 4 °C on a nutator.
- Under the fume hood, remove the 4% PFA fixative from the microcentrifuge tubes without touching the embryos. Discard the PFA in an appropriate container.
- Wash the embryos 5x in cold PBT for 10 min each at room temperature (RT).
- Using fine forceps, remove the heart and transfer it to a new 1.5 mL microcentrifuge tube with fresh PBT.
2. Whole mount immunofluorescence of the mouse embryonic heart
- Replace PBT with PBSTr (0.4% Triton X-100 in PBS) and wash the samples 3x, 5 min each, at RT on an orbital shaker.
- Replace PBSTr with blocking solution (PBSTr with 10% heat-inactivated bovine serum). Incubate from 2 h to O/N at 4 °C on an orbital shaker.
- Replace blocking solution with blocking solution containing primary antibody at the desired concentration (for anti-GFP or anti-VE-Cadherin antibody,use 1:1000 dilution). Incubate O/N on an orbital shaker at 4 °C.
- After O/N incubation at 4 °C, incubate for 1.5 h at RT on an orbital shaker. This step is very important to increase the signal-to-noise ratio.
- Remove and keep at 4 °C the primary antibody solution for up to three future experiments (add sodium azide to a final concentration of 0.02% for best conservation).
- Wash the embryos 3x with PBSTr for 3 min each.
- Wash the embryos with PBSTr 3x for 30 min each on an orbital shaker at 4 °C.
- After the last wash, add 1 mL of blocking solution containing 1:1000 fluorescence-conjugated secondary antibody against the primary antibody host species. Add also 1:3000 DAPI to the solution and incubate the embryos O/N on an orbital shaker at 4 °C.
- After O/N incubation at 4 °C, incubate for 1.5 h at RT on an orbital shaker. This step is very important to increase the signal-to-noise ratio.
- Wash the embryos 3x with PBSTr for 3 min each.
- Wash the embryos with PBSTr 3x-5x for 30 min each on an orbital shaker at RT.
3. Mounting the mouse embryonic AVC or OFTs
- Put the hearts on a Petri dish (35 mm) containing PBS.
- Using a tungsten wire and fine forceps, isolate the AVC or the OFT, and cut them longitudinally26.
- Prepare coverslips (22 mm x 22 mm), slides (60 mm x 24 mm), forceps, a P200 pipette with the appropriate tips, paper towel, and any aqueous glycerol-based mounting media for fluorescence, without DAPI.
- Using a similar approach as in Xiao, C. et al.25, stick two strips of tape on a slide, separated by 0.3-0.6 cm in order to create a 3D room space that will allow the samples to conserve it original shape without squashing them.
- Carefully, and using the minimum volume of buffer, drop the samples onto the slide between the tape stripes using a cut P200 tip pipette. Use a dissecting microscope to increase the accuracy.
- Using fine forceps, place the samples with the endocardium facing up (myocardium down on the slide).
- Remove excess PBS with a paper towel (avoid touching the sample), and then dry the samples for 1-2 min at RT to make the samples stick to the slide.
- Add 40 µL of aqueous-based mounting media on the tissue.
- Place the coverslip over the two pieces of tape and lower it slowly onto the tissue with a needle or forceps. Avoid creating air bubbles.
- Seal the coverslip using one drop of nail polish on each vertex of the coverslip.
- Once the nail polish is dry, clean the excess mounting media from the sides of the coverslip by using an absorbent paper towel. Avoid moving the coverslip.
- Add nail polish along the coverslip edges to fully seal it, preventing mounting media evaporation.
- Using an upright or an inverted confocal microscope, take images at 10x for general view and at 63x with 3x zoom for detailed images.
Representative Results
The data generated using this protocol show that it is possible to perform en face imaging of the endocardium of the AVC. The first objective was to analyze the cell shape of the endocardium during the formation of the valves at a cellular resolution (Figure 1). In order to highlight individual endocardial cells at E9.5, we used two transgenic mouse strains. (1) ROSAmT/mG is a two-color fluorescent Cre reporter allele (tdTomato/mT and EGFP/mG) whose fluorescence is detected at the cell membrane. Without Cre-mediated activation, the transgene expresses tdTomato (mT). Upon Cre-mediated activation, tdTomato expression is replaced by EGFP fluorescence expression (mG) in specific cells and clonal derivatives27. (2) The Cdh5CreERT2/+ mouse line expresses tamoxifen-inducible CRE-recombinase (CreERT2) under the regulation of the vascular endothelial cadherin promoter (Cdh5), which is expressed in the vascular endothelium and endocardium in the embryonic stages28. We crossed a male and a female of each genotype to obtain embryos carrying the two transgenes in order to activate the expression of GFP in individual cells of the endocardium. To achieve this, we inoculated low dosages of tamoxifen to activate CRE expression in a reduced number of cells. As a result, single cells, or clones of few cells, expressed GFP in the endocardium (Figure 1A). GFP expression (Figure 1A,B) in combination with VE-Cadherin subcellular localization (Figure 1C) is an effective approach to study the relationship between AJ dynamics and filopodia formation in pre-EMT cells since these cells develop membrane protrusions as AJs dissolve29. PCP signals can confer anisotropic contractility on the AJs, producing cellular forces that promote polarized morphogenesis in the epithelia30. We found differences in the intensity of VE-Cadherin staining in the endocardium (Figure 2), indicating that we cannot discard anisotropic contractility in the embryonic endocardium of AVC at pre-valvular stages.
The second objective was to analyze the planar topology of the endocardium during valve development (Figure 2). In order to define the number of cells converging in a single vertex, we stained the whole heart with VE-Cadherin antibody at E8.5 and E9.5 (Figure 2A,B). Other molecules localized at cell-cell contacts of the endocardial cell would also be useful for that purpose (e.g., β-Catenin). At E8.5, the endocardium of the AVC tends to have a stable epithelial organization, and we could not detect vertices formed by more than four cells (Figure 2A,C). Later at E9.5, we found vertices made up of up to six cells forming rosette-like structures (Figure 2B,D), similar to the cellular organization previously described in epithelia undergoing active cell rearrangements31, suggesting that the AVC endocardium is undergoing active cellular rearrangement during valve development.
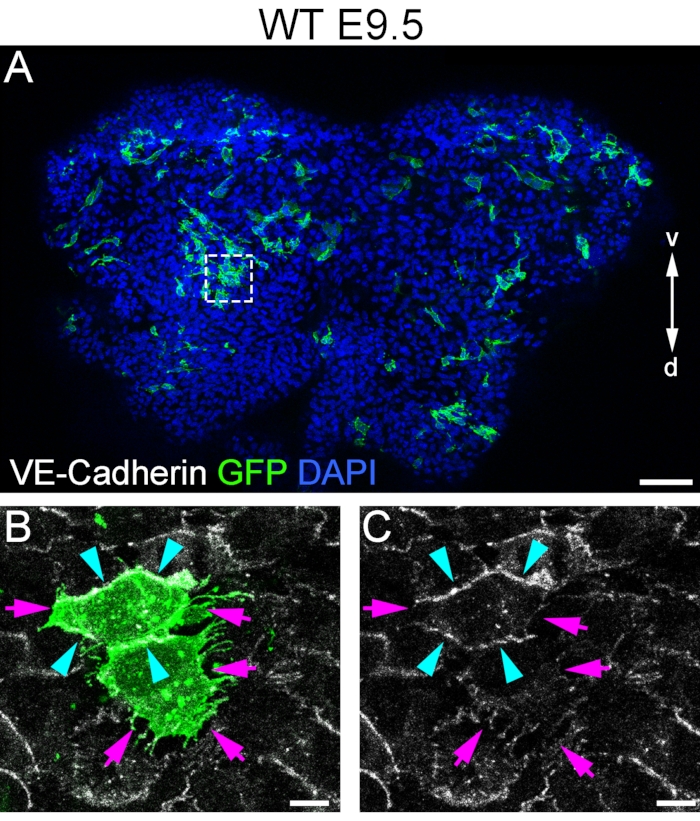
Figure 1: Single-cell shape analysis in prevalvular endocardium. (A) AVC from E9.5 mouse embryo opened longitudinally showing scattered GFP positive cells. (B) GFP expression in single cells defines cell shape at cellular resolution. (C) Whole-mount staining of VE-Cadherin highlights AJs. Filopodia are generated in domains of the cell that do not localize VE-Cadherin (purple arrows), and vice versa, VE-Cadherin localizes in domains of the cell that are smooth and do not form filopodia (blue arrowheads). v, ventral; d, dorsal. Scale bar in A is 100 µm; in B and C is 10 µm. Please click here to view a larger version of this figure.
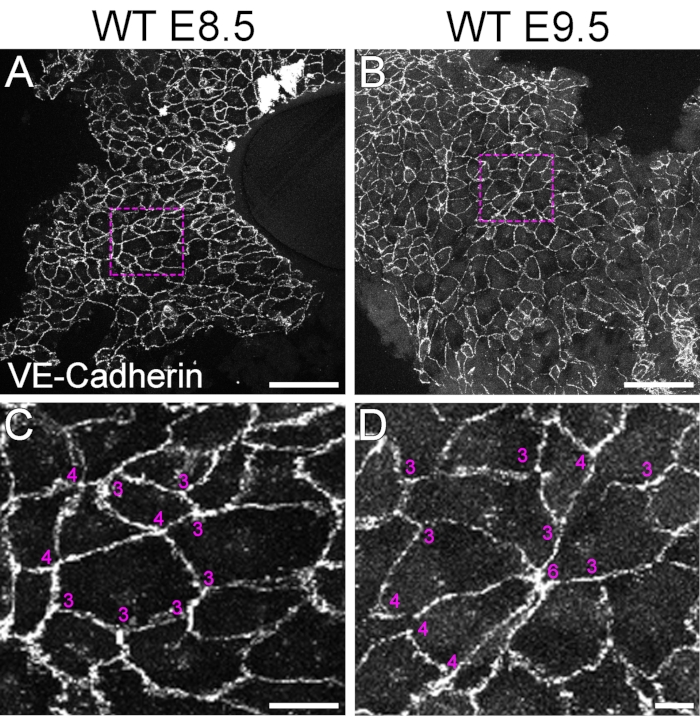
Figure 2: Planar topology and anisotropy of planarly localized proteins can be defined in en face images of endocardium at the AV canal. VE-Cadherin is localized at the cell-cell contacts of the endocardium. (A) E8.5 and (B) E9.5 AVC from embryos were opened longitudinally. Magnifications in (C) and (D) allowed us to observe the number of cells that converge in a vertex. In addition, we can distinguish different intensities in the VE-Cadherin staining, indicating that its location in the AVC is anisotropic. Scale bar in A and B is 100 µm; in C and D is 10 µm. Please click here to view a larger version of this figure.
Discussion
The endocardium is an epithelial monolayer that covers the entire inner surface of the embryonic heart tube. During valve development, endocardial cells at the prospective valvular regions undergo EMT, thus endocardial cells transform and rearrange their cytoskeleton to delaminate from the endocardium toward the cardiac jelly. We and others have obtained relevant data about valve development in mouse embryos by analyzing transversal sections of E8.5 and E9.5 embryonic hearts, where the endocardium is shown as a row of cells6,8,24,32. This approach allowed the analysis of the expression of signaling pathways and other molecules involved in valve development, but it was not possible to analyze spatial aspects of the pre-valvular endocardium.
AVC or OFT explants were mainly used to study transforming EC on a 3D collagen gel. Explants are useful for EMT quantification, the analysis of transformation capacity, or testing drugs after few days in culture6,7,11,33. These ex vivo experiments also allow the expansion of the endocardium on top of the collagen gel as a monolayer, but the environmental conditions in which ex vivo cardiac explants are grown differ from in utero conditions. For example, unidirectional blood flow is essential for proper valve development34,35 and is only achieved in in utero, not in ex vivo, explants.
This novel approach allows us to observe the intact valvular endocardium en face during the onset of valve development without any environmental manipulation. It allows analysis of the cellular distribution of the valvular endocardium, as well as single-cell shape analysis before and during EMT in in utero conditions. We can also observe and analyze the intensity and organization of the cell-cell contacts and the subcellular distribution of relevant molecules during EMT. This technique also offers the possibility of analyzing subcellular structures of the endocardium, the activation of signaling pathways by using reporter genes, the effect that gene inactivation may have on cellular behavior, and how it may affect wild-type neighboring cells.
To obtain high-quality samples, it is important to be very careful when handling them, since impurities (e.g., microfibers) that stick to the tissue can result in artifacts during the capture of images. In addition, as they are very small samples, the risk of breaking is high, so one must wash the tissue (step 1. and step 2.) in slow shakers. During the change of media with pipettes, we recommend cutting off the tip to prevent the tissue from collapsing as it passes through the tip in the event that it is accidentally absorbed. To become an expert in this technique, a learning curve is necessary, which will be longer or shorter depending on the researcher’s previous experience in micromanipulation.
Choosing the right fixative and time of fixation can be important in the case of the use of transgenes that express fluorescent proteins. The anatomical structure is not affected by different fixatives such as ethanol 70%, methanol, PFA 4%, or formalin 10%.
For counterstaining of nuclei, we recommend the incubation of samples with DAPI because penetration is much more efficient than using mounting media with DAPI.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This study was supported by grants PID2019-104776RB-I00 and CB16/11/00399 (CIBER CV) from MCIN/AEI/10.13039/501100011033 to J. L. P. J.G.-B. was funded by Programa de Atracción de Talento from Comunidad de Madrid (2020-5ª/BMD-19729). T.G.-C. was funded by Ayudas para la Formación de Profesorado Universitario (FPU18/01054). We thank the CNIC Unit of Microscopy and Dynamic Imaging, CNIC, ICTS-ReDib, co-financed by MCIN/AEI /10.13039/501100011033 and FEDER "A way to make Europe" (#ICTS-2018-04-CNIC-16). We also thank A. Galicia and L. Méndez for mouse husbandry. The cost of this publication was supported in part by funds from the European Regional Development Fund. The CNIC is supported by the ISCIII, the MCIN, and the Pro CNIC Foundation and is a Severo Ochoa Center of Excellence (grant CEX2020-001041-S) financed by MCIN/AEI /10.13039/501100011033.
Materials
| 4-OH-Tamoxifen | Sigma Aldrich | H-6278 | |
| 16 % Paraformaldheyde | Electron Microscopy Sciences | 157-10 | Dilute to 4% in water |
| anti-GFP | Aves Labs | FGP-1010 | |
| anti-VECadherin | BD Biosciences | 555289 | |
| Goat anti-Chicken, Alexa Fluor 488 | Thermo Fisher Scientific | A-11039 | |
| Goat Anti-Mouse Alexa Fluor 647 | Jackson ImmunoResearch | 115-605-174 | |
| DAPI | AppliChem | A4099,0005 | |
| Slides Superfrost PLUS | VWR | 631-0108 | 25 mm x 75 mm x 1.0 mm |
| Triton X-100 | Sigma Aldrich | X100-100ML | |
| Tween 20 | A4974,0500 | AppliChem | |
| Vectashield Mounting Medium | Vector Laboratories | H-1000-10 |
References
- Buckingham, M., Meilhac, S., Zaffran, S. Building the mammalian heart from two sources of myocardial cells. Nature Reviews Genetics. 6 (11), 826-835 (2005).
- Kelly, R. G., Buckingham, M. E., Moorman, A. F. Heart fields and cardiac morphogenesis. Cold Spring Harbour Perspectives in Medicine. 4 (10), 015750 (2014).
- Ivanovitch, K., et al. outflow tract heart progenitors arise from spatially and molecularly distinct regions of the primitive streak. PLoS Biology. 19 (5), 3001200 (2021).
- Rochais, F., Mesbah, K., Kelly, R. G. Signaling pathways controlling second heart field development. Circulation Research. 104 (8), 933-942 (2009).
- Moorman, A. F., Christoffels, V. M. Cardiac chamber formation: Development, genes, and evolution. Physiological Reviews. 83 (4), 1223-1267 (2003).
- Timmerman, L. A., et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes & Development. 18 (1), 99-115 (2004).
- Luna-Zurita, L., et al. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. Journal of Clinical Investigation. 120 (10), 3493-3507 (2010).
- Papoutsi, T., Luna-Zurita, L., Prados, B., Zaffran, S., de la Pompa, J. L. Bmp2 and Notch cooperate to pattern the embryonic endocardium. Development. 145 (13), (2018).
- MacGrogan, D., Luna-Zurita, L., de la Pompa, J. L. Notch signaling in cardiac valve development and disease. Birth Defects Research Part A: Clinical and Molecular Teratology. 91 (6), 449-459 (2011).
- de la Pompa, J. L., Epstein, J. A. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Developmental Cell. 22 (2), 244-254 (2012).
- Runyan, R. B., Markwald, R. R. Invasion of mesenchyme into three-dimensional collagen gels: a regional and temporal analysis of interaction in embryonic heart tissue. Developmental Cell. 95 (1), 108-114 (1983).
- Wu, B., et al. Nfatc1 coordinates valve endocardial cell lineage development required for heart valve formation. Circulation Research. 109 (2), 183-192 (2011).
- Amack, J. D. Cellular dynamics of EMT: Lessons from live in vivo imaging of embryonic development. Cell Communication and Signaling. 19 (1), 79 (2021).
- Cano, A., et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nature Cell Biology. 2 (2), 76-83 (2000).
- Batlle, E., et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nature Cell Biology. 2 (2), 84-89 (2000).
- Weng, M., Wieschaus, E. Myosin-dependent remodeling of adherens junctions protects junctions from Snail-dependent disassembly. Journal of Cell Biology. 212 (2), 219-229 (2016).
- Weng, M., Wieschaus, E. Polarity protein Par3/Bazooka follows myosin-dependent junction repositioning. Developmental Biology. 422 (2), 125-134 (2017).
- Jimenez-Amilburu, V., et al. In vivo visualization of cardiomyocyte apicobasal polarity reveals epithelial to mesenchymal-like transition during cardiac trabeculation. Cell Reports. 17 (10), 2687-2699 (2016).
- Davey, C. F., Moens, C. B. Planar cell polarity in moving cells: Think globally, act locally. Development. 144 (2), 187-200 (2017).
- Grego-Bessa, J., et al. The tumor suppressor PTEN and the PDK1 kinase regulate formation of the columnar neural epithelium. Elife. 5, 12034 (2016).
- Jones, C., Chen, P. Planar cell polarity signaling in vertebrates. Bioessays. 29 (2), 120-132 (2007).
- Mahaffey, J. P., Grego-Bessa, J., Liem, K. F., Anderson, K. V. Cofilin and Vangl2 cooperate in the initiation of planar cell polarity in the mouse embryo. Development. 140 (6), 1262-1271 (2013).
- Devenport, D. Tissue morphodynamics: Translating planar polarity cues into polarized cell behaviors. Seminars in Cell and Developmental Biology. 55, 99-110 (2016).
- Del Monte, G., Grego-Bessa, J., Gonzalez-Rajal, A., Bolos, V., De La Pompa, J. L. Monitoring Notch1 activity in development: Evidence for a feedback regulatory loop. Developmental Dynamics. 236 (9), 2594-2614 (2007).
- Xiao, C., Nitsche, F., Bazzi, H. Visualizing the node and notochordal plate in gastrulating mouse embryos using scanning electron microscopy and whole mount immunofluorescence. Journal of Visualized Experiments. (141), e58321 (2018).
- Mahler, G., Gould, R., Butcher, J. Isolation and culture of avian embryonic valvular progenitor cells. Journal of Visualized Experiments. (44), e2159 (2010).
- Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L., Luo, L. A global double-fluorescent Cre reporter mouse. Genesis. 45 (9), 593-605 (2007).
- Wang, Y., et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 465 (7297), 483-486 (2010).
- Yilmaz, M., Christofori, G. EMT, the cytoskeleton, and cancer cell invasion. Cancer and Metastasis Reviews. 28 (1-2), 15-33 (2009).
- Nishimura, T., Honda, H., Takeichi, M. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell. 149 (5), 1084-1097 (2012).
- Blankenship, J. T., Backovic, S. T., Sanny, J. S., Weitz, O., Zallen, J. A. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Developmental Cell. 11 (4), 459-470 (2006).
- Prados, B., et al. Myocardial Bmp2 gain causes ectopic EMT and promotes cardiomyocyte proliferation and immaturity. Cell Death and Disease. 9 (3), 399 (2018).
- Camenisch, T. D., Biesterfeldt, J., Brehm-Gibson, T., Bradley, J., McDonald, J. A. Regulation of cardiac cushion development by hyaluronan. Experimental & Clinical Cardiology. 6 (1), 4-10 (2001).
- Courchaine, K., Rykiel, G., Rugonyi, S. Influence of blood flow on cardiac development. Progress in Biophysics and Molecular Biology. 137, 95-110 (2018).
- Goddard, L. M., et al. Hemodynamic forces sculpt developing heart valves through a KLF2-WNT9B paracrine signaling axis. Developmental Cell. 43 (3), 274-289 (2017).

