An Intestine/Liver Microphysiological System for Drug Pharmacokinetic and Toxicological Assessment
Summary
We exposed a microphysiological system (MPS) with intestine and liver organoids to acetaminophen (APAP). This article describes the methods for organoid production and APAP pharmacokinetic and toxicological property assessments in the MPS. It also describes the tissue functionality analyses necessary to validate the results.
Abstract
The recently introduced microphysiological systems (MPS) cultivating human organoids are expected to perform better than animals in the preclinical tests phase of drug developing process because they are genetically human and recapitulate the interplay among tissues. In this study, the human intestinal barrier (emulated by a co-culture of Caco-2 and HT-29 cells) and the liver equivalent (emulated by spheroids made of differentiated HepaRG cells and human hepatic stellate cells) were integrated into a two-organ chip (2-OC) microfluidic device to assess some acetaminophen (APAP) pharmacokinetic (PK) and toxicological properties. The MPS had three assemblies: Intestine only 2-OC, Liver only 2-OC, and Intestine/Liver 2-OC with the same media perfusing both organoids. For PK assessments, we dosed the APAP in the media at preset timepoints after administering it either over the intestinal barrier (emulating the oral route) or in the media (emulating the intravenous route), at 12 µM and 2 µM respectively. The media samples were analyzed by reversed-phase high-pressure liquid chromatography (HPLC). Organoids were analyzed for gene expression, for TEER values, for protein expression and activity, and then collected, fixed, and submitted to a set of morphological evaluations. The MTT technique performed well in assessing the organoid viability, but the high content analyses (HCA) were able to detect very early toxic events in response to APAP treatment. We verified that the media flow does not significantly affect the APAP absorption whereas it significantly improves the liver equivalent functionality. The APAP human intestinal absorption and hepatic metabolism could be emulated in the MPS. The association between MPS data and in silico modeling has great potential to improve the predictability of the in vitro methods and provide better accuracy than animal models in pharmacokinetic and toxicological studies.
Introduction
Due to genomic and proteomic differences, animal models have limited predictive value for several human outcomes. Moreover, they are time-consuming, expensive and ethically questionable1. MPS is a relatively new technology that aims at improving the predictive power and reduce the costs and time spent with pre-clinical tests. They are microfluidic devices cultivating organoids (artificial mimetics functional units of organs) under media flow that promotes organoid-organoid communication. Organoids made of human cells increase translational relevance2,3,4. MPS is expected to perform better than the animal tests because they are genetically human and recapitulate the interplay among tissues. When fully functional, the MPS will provide more meaningful results, at higher speed and lower costs and risks4. Many groups are developing MPS for several purposes, especially disease models to tests drug’s efficacy.
Exposure level is one of the most critical parameters for evaluating drug efficacy and toxicity5,6,7,8,9,10,11,12. MPS allows organoid integration that emulates systemic exposure and is expected to perform better than the traditional 2D human tissue culture. This technology can significantly improve the prediction of compound intestinal absorption and liver metabolism4.
An MPS integrating human equivalent model of intestine and liver is a good starting point, considering the central role of these two organs in drug bioavailability and systemic exposure13,14,15. APAP is an attractive drug for studying an MPS without a kidney equivalent because its metabolization is done mainly by the liver16,17.
The 2-OC is a two-chamber microfluidic device suitable for the culture of two different human equivalent tissues/organoids interconnected by microchannels16. In order to emulate an in vitro human oral/intravenous administration of a drug and assess the effects of the cross-talk between the intestine and liver equivalents on APAP pharmacokinetics, besides the organoids functionality and viability, three different MPS assemblies were performed: (1) an “Intestine 2-OC MPS” comprised of an intestine equivalent based in a culture insert containing a Caco-2 + HT-29 cells coculture, integrated into the 2-OC device; (2) a “Liver 2-OC MPS” comprised of liver spheroids made of HepaRG + HHSteC (Human Hepatic Stellate Cells) integrated in the 2-OC device; and (3) an “Intestine/Liver 2-OC MPS” comprised of the intestine equivalent in one device compartment communicating with the liver equivalent in the other by the media flow through the microfluidic channels.
All assays were performed under static (no flow) and dynamic (with flow) conditions due to the impact of the mechanical stimuli (compression, stretching, and shear) on the cell viability and functionalities18,19,20. The present article describes the protocol for APAP oral/intravenous administration emulation and the respective absorption/metabolism and toxicological analyses in the 2-OC MPS containing human intestine and liver equivalent models.
Protocol
1. Production of tissue equivalents for cultivation in the 2-OC
- Small intestine barrier equivalent production
- Maintain Caco-2 and HT-29 cells using the intestine equivalent medium: DMEM supplemented with 10% FBS, 1% penicillin and streptomycin, and 1% non-essential amino acids, which is named as “DMEM S” in this manuscript.
- Remove the medium, wash twice with 1x DPBS and add 8 mL of 0.25% Trypsin/EDTA to dissociate Caco-2 cells grown in cell culture flasks (175 cm2). Incubate for 5 min at 37 °C and stop the reaction by adding at least the double volume of trypsin inhibitor. Perform the same procedure for HT-29 cells, adjusting the reagent volumes since a smaller quantity of these cells is needed and they are maintained in smaller flasks (75 cm2).
- Centrifuge at 250 x g for 5 min, remove the supernatant from both tubes, and resuspend the cell pellets in 10 mL of DMEM S. Count cells, assuring a cell viability higher than 80%. Aseptically integrate cell culture inserts in a 24-well plate previously filled with 400 µL of DMEM S per well in the basolateral side (which represents the human bloodstream).
- Co-cultivate Caco-2 and HT-29 cells at a ratio of 9:121. Use 2.25 x 105 Caco-2 and 2.5 x 104 HT-29 cells to each intestine equivalent in a final volume of 200 µL of DMEM S. Adjust cell numbers and volume according to the desired number of organoids. Mix carefully.
- Pipette 200 µL of cell solution into each insert’s apical side (which represents the human intestinal lumen side), seeding 250,000 cells per insert. Co-cultivate the cells in the inserts for three weeks22. Change the medium at least three times a week, aspirating it from both the apical and basolateral sides with a sterile Pasteur pipette, taking care not to damage the intact cell barrier.
NOTE: Proceed with the aspiration on the apical side, so as not to touch the cell barrier (aspirate by supporting the Pasteur pipette on the plastic rim of the cell insert). - Check the tight monolayer formation by measuring the TEER (transepithelial electrical resistance) every three days using a voltmeter23, according to the manufacturer's instructions.
- Perform a blank, measuring the resistance across a cell culture insert without cells, but with the same cell medium and at the same cell plate.
- Calculate tissue resistance by subtracting the blank resistance from the tissue-equivalent resistance, and multiply by the effective surface area of the filter membrane (0.6 cm2). A good intestine barrier resistance is in a range of 150 to 400 Ω∙cm2.
NOTE: After 21 days the cells must be fully differentiated and the intestinal barrier formed, so the intestine equivalents are ready to be integrated into MPS.
- Liver equivalents production
- Maintain HepaRG cells using the liver equivalent medium, which is William’s Medium E supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 units/mL penicillin, 100 µg/mL streptomycin, 5 µg/mL human insulin and 5 x 10-5 M hydrocortisone, and is named “Williams E S” in this manuscript. Renew HepaRG media every 2-3 days and maintain cell culture for two weeks to initiate the differentiation in hepatocytes and cholangiocytes.
- After the first two weeks, add 2% DMSO to HepaRG’s medium for an additional two weeks to complete the cell differentiation24,25. Grow HHSTeC in Stellate Cell Media (SteC CM), using poly-L-lysine-coated cell culture flasks, changing media every two or three days.
- Remove the medium, wash twice with 1x DPBS and add 8 mL of 0.05% Trypsin/EDTA, to dissociate HepaRG cells grown in cell culture flasks (175 cm2). Incubate for 5 to 10 min at 37 °C and stop the reaction by adding at least double the volume of trypsin inhibitor. Perform the same for HHSTeC, adapting the reagent volume since a smaller quantity of these cells are needed and they can be maintained in smaller flasks (75 cm2).
- Centrifuge both at 250 x g for 5 min, remove the supernatant and resuspend the cell pellets in Williams E S medium. Count cells, assuring a cell viability higher than 80%.
- Generate the liver spheroids combining HepaRG and HHSTeC cells at a ratio of 24:1, respectively, in Williams E S medium16. Add 4.8 x 104 differentiated HepaRG and 0.2 x 104 HHSTeC to compose each liver spheroid of 50,000 cells, in a volume of 80 µL. Adjust cell numbers and volume according to the desired number of spheroids. Mix carefully.
- Using a multichannel pipette, dispense 80 µL of the combined cell pool in each well of 384 spheroid microplates, which has round well-bottom geometry.
NOTE: After four days, spheroids of about 300 µm are formed. - Using wide-bore tips, transfer the liver spheroids to ultra-low-attachment 6 well plates, which allows the required “one-by-one” counting.
2. Integration of intestine and liver equivalents in a 2-OC MPS
- Intestine 2-OC MPS assembly for absorption assay
- Pipette 500 µL of DMEM S into the larger compartment of 2-OC and 300 µL in the smaller one. Aspirate the basolateral and apical media of each intestinal barrier equivalent in the 24 well plates. Using sterile forceps, integrate one insert per 2-OC circuit, specifically into the larger compartment. Apply 200 µL of the intestine medium at the apical side.
NOTE: Avoid the formation of bubbles when integrating the organoids into the MPS. - Connect the MPS to the control unit, which must be connected to a pressurized air supply. Set the parameters: a pressure of, approximately, ±300 bar and a pumping frequency of 0.3 Hz. Start the flow 24 h before the test substance administration. The next day, perform the APAP treatment.
- Pipette 500 µL of DMEM S into the larger compartment of 2-OC and 300 µL in the smaller one. Aspirate the basolateral and apical media of each intestinal barrier equivalent in the 24 well plates. Using sterile forceps, integrate one insert per 2-OC circuit, specifically into the larger compartment. Apply 200 µL of the intestine medium at the apical side.
- Liver 2-OC MPS assembly for metabolism assay
- Pipette 650 µL of Williams E S into the large compartment and 350 µL into the smaller compartment, which will receive the spheroids. In the ultra-low-attachment 6 well plates, count the spheroids using wide-bore tips. Each liver equivalent is composed of twenty spheroids26. Integrate twenty liver equivalents per circuit, using wide-bore tips, which permits the transfer of organoids only, into the smaller compartment of a 2-OC.
- Connect the MPS to the control unit, which must be connected to a pressurized air supply. Set the parameters: a pressure of, approximately, ±300 bar and a pumping frequency of 0.3 Hz. Start the flow 24 h before the test substance administration. The next day, perform the APAP treatment.
- Intestine/Liver 2-OC MPS assembly for absorption and metabolism assay
- Combine the two media (intestine and liver) in a 1:4 proportion, which means 200 µL of DMEM S in the intestine apical side and 800 µL of Williams E S in the basolateral side. Integrate intestine and liver equivalents, simultaneously, in the 2-OC.
- Connect the MPS to the control unit, which must be connected to a pressurized air supply. Set the parameters: a pressure of, approximately, ±300 bar and a pumping frequency of 0.3 Hz. Start the flow 24 h before the test substance administration. The next day, perform the APAP treatment.
NOTE: For all experiments, perform each time point in triplicate, which means three separated 2-OC circuits (i.e., 1 and ½ 2-OC devices). The total volume of each 2-OC circuits is 1 mL.
3. Acetaminophen (APAP) preparation
- Prepare the APAP stock solution, dissolving APAP in absolute ethanol. On the day of the experiment, dilute APAP in the respective medium (APAP solution), to a concentration of 12 µM for “oral administration” and 2 µM for “intravenous administration”.
- Ensure that the final concentration of ethanol in the vehicle control and treatment solution is 0.5% for both administrations. For the positive control (100 mM APAP), the ethanol concentration is 2%.
4. Test substance administration and media sampling
- APAP “oral” administration and media sampling
- Aspirate the basolateral and apical media of each intestinal barrier equivalent in the 2-OC. Pipette 500 µL of the appropriate culture medium into the large compartment at the organoid basolateral side and 300 µL into the small compartment.
- Check for bubbles and proceed with the intestinal barrier equivalent treatment with the test substance in the apical side, emulating oral administration. Emulate APAP “oral” administration by adding 200 µL of a 12 µM APAP solution on the apical side of intestinal culture inserts, which represents the intestinal “lumen side” (Figure 1B). Connect the MPS to the control unit.
- Collect the total volume from apical and from basolateral sides at the following time points: 0 h, 5 min, 15 min, 30 min, 1 h, 3 h, 6 h, 12 h, and 24 h15,27. Perform all experiments in triplicate, at static and dynamic conditions, and collect each sample, of each triplicate, in a separate microtube. Analyze the samples using HPLC/UV.
NOTE: Separate apical and basolateral samples.
- APAP “intravenous” administration and media sampling
- Emulate the “intravenous” route by administering 2 µM APAP solution directly into the liver compartment. Aspirate all 2-OC medium content. Pipette 650 µL of Williams E S containing the test substance into the large compartment and 350 µL of the same media into the smaller compartment which contains the 20 spheroids. Collect all volumes at the following time points: 0 h, 30 min, 1 h, 2 h, 3 h, 6 h, 12 h, and 24 h27,28.
- Perform all experiments in triplicate, at static and dynamic conditions. Collect each sample, of each triplicate, in a separate microtube. Analyze the samples using HPLC/UV.
5. Instrumentation and chromatographic conditions
- HPLC analysis
- Set all relevant parameters for the HPLC analysis according to Table 1.
- Filter the mobile phase through a 0.45 µm membrane filter under vacuum. Filter the samples through a 0.22 µm pore size PVDF syringe filter (diameter 13 mm) and store them in a vial. Start the measurement.
- Stock solutions, calibration standards, and quality control (QC) samples
- Prepare 10 mM of APAP stock solutions in ammonium acetate buffer (100 mM, pH 6.8) and further dilute with DMEM S and Williams E S cell culture media diluted with ammonium acetate buffer (1:1, v/v) to achieve the working solutions ranging from 0.25 to 100.00 µM.
- Include a set of calibration samples in triplicate as well as quality control samples at four levels in triplicate. Prepare these standards by serial dilution.
- Create calibration curves of APAP peak areas versus APAP nominal standard concentrations. Determine the linear regression fit for each calibration curve. Evaluate the goodness-of-fit of various calibration models by visual inspection, correlation coefficient, intra- and inter-run accuracy and precision values.
- Inject blank samples of DMEM S and Williams E S media diluted in ammonium acetate buffer (1:1, v/v) in sextuplicate. Prepare triplicates of quality control samples in DMEM S and Williams E S media diluted with ammonium acetate buffer (1:1, v/v) for the APAP concentrations of 0.50 (LOQ), 4.50, 45.00 and 90.00 µM.
- Ensure that the quality control samples are prepared from a new stock solution, different from that used to generate a standard curve. Use quality control samples to investigate intra- and inter-run variations.
- Validation procedures
- Perform the bioanalytical method validation following the previously reported procedures29,30. Carry out the chromatographic runs on five or six separate occasions, considering Williams E S and DMEM S cell culture media, respectively.
- Ensure that calibration points ranging from 0.25 to 100.00 µM of APAP, in DMEM S or Williams E S cell culture media diluted in ammonium acetate buffer (1:1, v/v), are plotted based on the peak-areas of APAP (axis y) against the respective nominal concentrations (axis x). Compare the slopes of these standard calibration curves with slopes of calibration curve prepared in ammonium acetate buffer. Make sure that all calibration curves have a correlation value of at least 0.998.
- Determine the precision and accuracy (intra and inter-run) for the analyte in the surrogate matrix using replicates at four different levels LLOQ, low, middle, and high-quality control on five or six different days. Perform intra-run precision and accuracy measurements on the same day in DMEM S or Williams E S cell culture media diluted in ammonium acetate buffer (1:1, v/v) containing 0.50, 4.50, 45.00 and 90.00 µM APAP concentrations (n= 3).
- Evaluate each set of quality control samples containing the APAP concentrations from recently obtained calibration curves. Test the selectivity of the assays by the degree of separation of the compound of interest and possible other chromatographic peaks caused by interfering components.
- Lower limit of quantification (LLOQ) and limit of detection (LOD)
- Determine the lower limit of quantification (LLOQ) based on the standard deviation of the response and the slope approach. Calculate using the formula 10α/S, where α is the standard deviation of y-intercept and S is the slope of straight line obtained by plotting calibration curves29,30. Estimate the limit of detection (LOD) taking into consideration 3.3 times the standard deviation of the blank, divided by the slope of the calibration curve29,30.
6. Tissue equivalents viability/functionality
- MTT
- Perform an MTT assay to assess organoid viability in all time points of the MPS assay. As the negative control, use cell media plus vehicle. As the positive control, treat the organoids with 100 mM APAP and 1% NaOH diluted in cell medium.
- Transfer the 20 spheroids of each replicate for individual wells in a 96 well plate, and the cell culture inserts, containing the intestine equivalents, to 24 well cell plates, placing one intestine equivalent per well. Wash the tissue equivalents three times with 1x DPBS.
- Add 300 µL of a 1 mg/mL MTT solution, diluted in the respective cell medium, per well. Incubate the plates for 3 h at standard cell culture conditions.
- Remove the MTT solution from each well carefully by pipetting. Extract MTT formazan from the intestine and liver equivalents using 200 µL of isopropanol per well overnight at 4 °C.
NOTE: Seal the lid to prevent evaporation. - Transfer 200 µL of each supernatant to the respective pre-identified well in a 96 well micro test plate. Use isopropanol as the blank.
- Read the formazan absorbance in a plate reader at 570 nm. Calculate the relative ability of the cells to reduce MTT (%) using the average optical density of each time point, compared to the negative control, considered as 100% cell viability.
- Cytochemistry/Histology
- Fix the intestine and liver equivalents, for 25 min at room temperature, using 4% (w/v) paraformaldehyde in 0.1 M phosphate-saline buffer, pH 7.4. Wash the organoids 5 times in PBS buffer for 10 minutes each time. Stain the intestinal and the liver equivalents with tetramethylrhodamine isothiocyanate-phalloidin or Alexa Fluor 647 phalloidin, 1:50 in PBS31.
- Transfer them to OCT freezing medium for a few minutes to acclimate at RT before transferring them to liquid nitrogen until the complete freezing. Perform liver spheroids cryosections about 10-12 µm thick, using a cryostat.
- Mount the tissues sections in mounting medium with DAPI. Examine them by confocal fluorescence microscopy.
- Freeze the organoids after fixation to perform hematoxylin & eosin staining according to the established protocols. Mount the slides with mounting medium after slicing the tissue as described above and take histological images using an optical microscope.
- High content analysis
- Mitochondrial and nuclear staining of the cells
- Reconstitute the lyophilized powder in DMSO to make a 1 mM mitochondrial staining stock solution (e.g., MitoTracker Deep Red FM). Store aliquoted stock solution at -20 °C protected from light. Dilute the 1 mM mitochondrial staining stock solution to the final concentration (200 nM) in prewarmed (37 °C) tissue culture medium without serum.
- Remove the cell culture media. Add the mitochondrial staining solution to completely cover the sample and incubate cells for 15-45 min at 37 °C in a humidified atmosphere with 5% CO2.
- Carefully remove the mitochondrial staining working solution and replace it with 2-4% paraformaldehyde fixative in PBS for 15 minutes at room temperature.
- Rinse the fixed cells gently with PBS for 5 minutes. Repeat the washing process twice.
- Prepare a 10 mg/mL (16.23 M) nucleic acid staining stock solution by dissolving 100 mg of Hoechst 33342 dye in 10 mL of ultrapure water.
NOTE: The stock solution should be aliquoted and stored protected from light at -20 °C. - Prepare a 0.2-2.0 µg/mL nucleic acid staining working solution in PBS and incubate the fixated cells with nucleic acid staining working solution for 10 minutes at room temperature.
- Remove the nucleic acid staining working solution and rinse the cells gently with PBS for 5 minutes three times. Cells should be kept in PBS at 4 °C, protected from light.
- Mitochondrial and nuclear staining analysis
- Analyze cells using a fluorescence microscope with filter sets appropriate for the nucleic acid stain (λEx/ λEm: 361/497 nm) and the mitochondrial stain (λEx/ λEm: 644/665 nm). Find cells by nucleic acid positive staining and quantify cell number. Quantify mitochondrial stain fluorescence intensity in mitochondria.
- Mitochondrial and nuclear staining of the cells
- Morphometric measurements (spheroids calculations) in ImageJ
- Export High Content Analysis (HCA) images as *.flex files from the Columbus software. Import .flex files as a grayscale in ImageJ using Bio-Formats plugin32: File > Import > Bio-Formats.
- In the Import Options window, select Hyperstack viewing and enable Split channels under Split into separate windows. This option will allow the access of all files in a particular channel (e.g., DAPI, mitotracker, etc.). Do not select Use virtual stack under Memory management.
NOTE: It is preferable to use DAPI channel as “dust” in images as culture medium is reduced in UV wavelength (e.g., 405 nm). - Adjust pixel size (Analyze > Set Scale) if it was not loaded according to embedded values in the .flex file. Apply a Gaussian Blur filter to remove the excess of noise and avoid irregularities in shape contour. Process > Filters > Gaussian Blur. A high value of Sigma (radius) between 2.0 and 3.0 is ideal for most cases. If the stack has several images, apply to all of them (select Yes in Process Stack window).
- Generate a binary image to separate background and organoids (objects) using a threshold. Click Image > Adjust > Threshold. Use the red mask to adjust the values according to the intensity of the image, to fit the organoid shape, keeping the morphology intact. Disable Dark background if the image has a white background. Click Apply.
- In the Convert Stack to Binary window, choose the Threshold method. Usually, Default or Triangle are preferred in this kind of image processing. Keep the Background as Dark. Select Calculate threshold for each image if there are several images in the stack.
- Select Process > Binary > Fill holes. Optionally, remove holes from the background. In Process > Binary > Options, select Black background and execute Fill Holes again. Disable Black background option before proceeding to the next step.
- Separate objects. For organoids, the watershed method is a good choice. Click Process > Binary > Watershed. Execute the shape analysis.
- Select Analyze > Set Measurements. Several options are available (details in https://imagej.nih.gov/ij/docs/guide/146-30.html). For organoids, select Area, Mean gray value, Min & max gray value and Shape descriptors. Optionally, select Display label to identify objects in the image and Scientific notation. Click OK.
- Select Analysis > Analyze Particles. Choose the Size and Circularity limits. Keep 0-infinity and 0.00-1.00, respectively to measure all objects in the image. In Show, choose Outlines so the objects will be identified. Enable Display results to output results; Exclude on edges to exclude objects touching borders; Include holes so eventual interior holes in the objects are considered as part of the main shape.
- Repeat Shape Analysis for every image stack and the results will be appended in a single table. Export results table in File > Save As… as Comma Separated Values (.csv) file.
- Real-Time PCR
- Extract RNA from tissue equivalents using a monophasic solution of phenol and guanidine isothiocyanate, following manufacturer’s instructions.
- Perform the cDNA synthesis by reverse transcription of 1 – 2 µg of total RNA.
- Amplify all targets using gene-specific primers (Table 5) to perform real time quantitative PCR. Each qRT-PCR contains 30 ng of reverse-transcribed RNA and 100 nM of each primer.
- Follow PCR conditions: 50 °C for 3 minutes (1 cycle); 95 °C for 5 minutes (1 cycle); 95 °C for 30 seconds, 59 °C for 45 seconds and 72 °C for 45 seconds (35 – 40 cycles).
- CYP assay
- Follow section 2.2 for liver 2-OC assembly. The experimental groups are No-cell control, APAP 2 µM treatments for 12 h, 24 h, and vehicle control. Follow sections 3.3 and 4.2 for APAP 2 µM preparation and treatment.
NOTE: To ensure that all the samples will be ready at the same time for CYP assay, start the 12 h treatment 12 hours after starting the 24 h treatment. Treat the no-cell control of CYP activity with 0.5% ethanol solution, as well as the vehicle control. - Thaw 3 mM luminogenic substrate stock solution at room temperature and make a 1:1000 dilution in William’s E S. Protect from light.
- Collect spheroids and transfer each experimental group to a well of a 96-well plate. Remove the medium, wash twice with 100 µL of 1x DPBS and add 80 µL of 3 µM substrate solution per well. Keep the no-cell control in William’s E S medium. Save a well without spheroids or substrate solution for a background control. Incubate for 30-60 min at 37 °C with 5% CO2, protected from light.
- Equilibrate lyophilized Luciferin Detection Reagent (LDR) using the Reconstitution Buffer with esterase. Mix by swirling or inverting. Store the appropriated volume at room temperature until the next step.
NOTE: Reconstituted LDR can be stored at room temperature for 24 hours or at 4 °C for 1 week without loss of activity. For long-term storage, store at –20 °C. - Transfer 25 µL of intact spheroids’ supernatant in three different wells of a white opaque 96-well microplate, after incubation. Add 25 µL of LDR per well and homogenize.
- Incubate the white plate at room temperature for 20 minutes. Read the luminescence on a luminometer. Do not use a fluorometer.
- Calculate net signals by subtracting background luminescence values (no-cell control) from the test compound-treated and untreated (vehicle control) values. Calculate percent change of CYP3A4 activity by dividing the net treated values by the net untreated values and multiplying by 100.
- Follow section 2.2 for liver 2-OC assembly. The experimental groups are No-cell control, APAP 2 µM treatments for 12 h, 24 h, and vehicle control. Follow sections 3.3 and 4.2 for APAP 2 µM preparation and treatment.
- Western blotting
- Transfer the liver spheroids to an identified 1.5 mL microtube. Remove the medium and wash twice with 100 µL of 1x DPBS.
- Lysate the liver spheroids in 100 µL of cell lysis RIPA buffer at 4 °C for 20 min. Centrifuge for 15 min, 4 °C, and 11000 rpm. Transfer the supernatant to another identified 1.5 mL microtube.
- Quantify the amount of protein obtained through the Bradford method. Load between 10 and 50 µg of protein from the quantified cell lysate per well of a gradient polyacrylamide gel 3-15% and perform an SDS-PAGE.
- Transfer the loaded protein from the gel to a 0.22 µm PVDF membrane through the semi-dry system equipment. Use a transference solution of 50 mM Tris-HCl and 192 mM glycine. Set equipment parameters according to the number of gels to be transferred (1 to 2 at a time).
- Block nonspecific interactions on PVDF membrane with a 3-5% skim milk solution in TBS-T buffer: Tris-Buffered Saline (50 mM Tris pH 7.6, 150 mM sodium chloride) supplemented with 0.1% of Tween 20. Keep the membrane under gently constant shaking for 1 hour at room temperature.
- Wash with TBS-T under gently constant shaking for 3-5 min at room temperature. Repeat this washing step twice.
- Dilute albumin and vinculin primary antibodies to 1:1000 and 1:2000 respectively on TBS-T. Incubate the membrane with primary antibodies overnight at 4 °C, under gently constant shaking.
NOTE: Always follow the manufacturer’s instructions to dilute antibodies. - Remove the primary antibody and wash membrane 3 times (step 6.7.6). Dilute ECL anti-mouse IgG secondary antibody to 1:5000 on TBS-T. Incubate the membrane with a secondary antibody under gently constant shaking for 2 hours at room temperature.
- Remove the secondary antibody and wash the membrane (step 6.7.6). Perform protein detection using ECL Western Blotting Substrate. Expose the autoradiographic films for 30 s to 30 min. Perform the immunoblotting detection in triplicate.
Representative Results
To perform the PK APAP tests in the 2-OC MPS, the first step is to manufacture the human intestine and liver equivalents (organoids). They are integrated into the 2-OC microfluidic device (Figure 1A) 24 h before starting of the PK APAP assay. The next day, the medium is changed, and the model is exposed to APAP. Figure 1 illustrates the intestine and liver equivalents placed inside the 2-OC device (Figure 1B) and the APAP PK experiment time course (Figure 1C). We performed a MTT assay, TEER measurements, HCA, real-time PCR, western blotting, histology, and confocal fluorescence microscopy in 2D culture and 3D organoids to check the tissue’s viability and detect possible APAP toxic effects. In the confocal fluorescence microscopy images, the intestine equivalent samples stained with DAPI and Phalloidin (for nuclei and actin respectively) were presented as a contiguous barrier for the non-treated (Figure 2A) and the 12 µM APAP treated samples (Figure 2B). As seen in Figure 2C, the MTT assay showed relative cell viability levels above 70%, indicating the absence of relevant cytotoxic effects in response to APAP exposure at 12 µM concentration33,34,35,36,37. The positive control (100 mM) induced significant cell death (survival below 5%). The Caco-2/HT-29 viability and proper differentiation as well as the intestine equivalent barrier integrity were verified by TEER evolution during the differentiation period (Figure 2D). APAP did not cause any alteration in the TEER values as shown in Figure 2E. The expression of the active sodium-coupled glucose transporters SLC5A1, multidrug resistance transporter MDR1 and sodium-potassium ATPase were analyzed, to verify the APAP treatment impact over cell barriers formation and basal functionality. As demonstrated in Figure 2F–H, both non-treated and APAP treated intestine equivalents have shown similar expression of SLC5A1 and NaKATPase. The oral administration of 12 µM APAP induced marked an increase in the MDR1 mRNA levels in intestines equivalents after 24 h (Figure 2H). We also performed HCA of cell phenotypic changes by a fluorophore dye mixture for nuclei and mitochondrial mass content. Positive controls were 100 mM APAP and 1% NaOH.
Additionally, we analyzed whether 12 µM APAP could induce cytotoxicity to the 2D Caco-2/HT-29 co-culture. The intestinal cells images acquired with fluorescence microscopy shown in Figure 2I corroborates the MTT data, which have demonstrated that 12 µM APAP did not cause significant cytotoxicity in the Caco-2/HT-29 intestinal equivalents.
The assessment of hepatic spheroids basal viability and the cytotoxicity in response to 2 µM APAP were done by MTT assay and morphological analyses by confocal fluorescence microscopy, H&E histology, and HCA assays. As shown in Figure 3A, the MTT assay was unable to identify any relevant cytotoxicity in response to the 2 μm APA treatment in samples taken from the Liver 2-OC assembly under both static and dynamic conditions. The cell viability decreased but remained over 80% for both 12 h and 24 h time-points33,34,35,36,37. The positive control treatments (100 mM APAP and 1% NaOH) induced significant tissue damages (viability below 10%). Microscopic confocal images indicate the absence of a necrotic center in the liver spheroids in both basal or APAP treatment conditions, and no evidence of significant death rates (Figure 3B-C). However, when we analyzed multiple cellular phenotypic changes following the vehicle or 2 µM APAP administration in the 2D HepaRG/HHSteC coculture through HCA assay, using 100 mM APAP (C+) as a positive control, contradictorily to the results of the MTT assay, the hepatic cells demonstrated early cytotoxic responses to 2 µM APAP treatment (Figure 3D). After 24 h, there was a decrease in the number of cells, in the nuclear area and an increase in mitochondrial mass. Additionally, a fluorophore dye cocktail containing Hoechst 33342 and MitoTracker Deep Red was used to stain the 3D hepatic spheroids (Figure 3J). Fiji software was used to evaluate 3D spherical architecture homogeneity among several spheroids (Figure 3E–I). The graphic shown in Figure 3E shows the similarity among liver spheroids total area. The aspect ratio (Figure 3F) around 1 means an absence of bias during the confection process of the spheroids. The evaluation also indicated that the majority of the spheroids were roughly rounded (Figure 3G). The evaluation of the morphology perimeter and cell distribution was done by circularity (Figure 3I) and by solidity calculation (Figure 3H), respectively. We concluded that the methodology to confectioning the liver spheroids had generated organoids with a smooth perimeter, compatible with spherical growth, no biases, or necrosis during the process.
As demonstrated in Figure 4A-B, the liver spheroids showed a relative basal high level of albumin and GST mRNA expression respectively, indicating proper basal functionality. Nevertheless, the 2 µM APAP treatment for 24 h induced a decrease in the albumin and the GST mRNA expression levels, suggesting impairment of liver spheroids functionally at 24 h time point of APAP treatment.
The detection of the CYP3A4 and UGT1A1 mRNA expression levels demonstrates the liver equivalents metabolic capacity. The CYP3A4 mRNA basal level (Figure 4C) was consistent with previous reports6. The APAP treatment induced a trend for a decrease in both CYP3A4 mRNA and UGT1A1 expression in response to APAP treatment (Figure 4C-D) once again corroborating the hypothesis of impairment of liver spheroids functionally at 24 h time point of APAP treatment.
Additionally, experiments of western blotting and in vitro enzymatic activity were performed in order to analyze the albumin protein expression, as well as, the CYP 3A4 activity in liver equivalents at basal and at APAP treatment conditions. We found that 2 µM APAP treatment performed at the Liver 2-OC MPS, induced a reduction in the liver equivalent total albumin expression at 12 h and 24 h time-points at both static (Figure 4E) and dynamic (Figure 4F) conditions. On the other hand, liver equivalents samples from dynamic conditions demonstrated a trend to present higher levels of protein expression of albumin when compared to the static conditions (Figure 4G). CYP 3A4 in vitro assay performed at liver equivalents from Liver 2-OC MPS shown that 2 µM APAP treatment for 12 h or 24 h was capable to induce a robust and significant impairment of CYP 3A4 activity at both static and dynamic conditions (Figure 4H-I). More interesting, the presence of media flow (dynamic) has induced a significant improvement of liver equivalents CYP 3A4 activity levels when compared to conditions in which the liver equivalents were kept in absence of circulating medium (static conditions) (Figure 4J).
To find the most sensitive analytical condition regarding HPLC analysis, several parameters were investigated including the composition of the mobile phase, the type, and concentration of additives. It was found that acetonitrile gave better chromatogram resolution and appropriate retention time than methanol. Fast and reproducible separation of APAP was obtained using a C18 reversed-phase column. The APAP retention time (Rt) value was 9.27 ± 0.19 minutes. Selectivity for APAP is indicated by the shape and symmetrical resolution of the peak, as well as by the lack of interfering peaks from the DMEM and Williams cell culture media.
APAP standard concentrations in DMEM S and Williams E S cell culture media diluted with ammonium acetate buffer (1:1, v/v) ranging from 0.25 to 100.00 µM were used to build the calibration curves. The linearity of the method was determined at nine concentration levels. The data are shown in Table 2 and Table 3. The relationship between APAP concentration and the peak areas was described by the linear regression equations: y = 16106*x + 3579.8 (R2=1, in DMEM medium) and y = 16397*x + 2475.1 (R2=1, in Williams medium), in which “x” is APAP nominal concentration in µM and “y” is the chromatogram peak area of APAP in AU. At the upper limit of quantification (i.e., 100.00 µM), the percentage deviation and the inter-run variability values were less than 2.50%. The accuracy and the precision for nine concentration levels, excluding the 0.50 µM (LLOQ), were within an acceptable range with DEV and C.V. values less than 7.00% (Table 2 and Table 3).
The analytical method inter- and intra-run accuracy and precision, at four tested concentrations, fell within the generally accepted criteria for bioanalytical assays. The reproducibility of the method was evaluated by analyzing replicates of APAP quality control samples of 0.50 (LLOQ), 4.50, 45.00 and 90.00 µM. The intra-run and inter-run average results are reported in Table 4. The accuracy and precision of the assay are demonstrated by DEV values ≤ 15.00% and by C.V. values ≤ 7.00%, respectively.
LOD was determined as the sample whose signal-to-noise ratio (S/N) was just greater than 3 and corresponded to a 0.25 µM APAP. On the other hand, the LLOQ, estimated with 0.50 µM APAP samples, displayed S/N ratio equal to 10. Furthermore, we found accuracy values (DEV%) ranging within ≤ 19.00% of the nominal concentration values. The intra- and inter-run variabilities were demonstrated by C.V. ≤ 18.77%, as shown in Table 2, Table 3 and Table 4)29,30.
The APAP PK analyses were performed in three different 2-OC MPS assemblies: 1) Intestine 2-OC, containing the intestine equivalent only; 2) Liver 2-OC, containing the liver spheroids only and 3) Intestine/Liver 2-OC with both intestinal barrier and liver spheroids.
For absorption studies, the oral route was mimicked by the administration of 12 µM APAP on the intestine equivalent apical side. APAP concentrations were measured by HPLC/UV, in the medium samples, collected from apical and basolateral intestinal equivalents sides, in both static and dynamic conditions. The APAP kinetics in the medium collected from the apical and basolateral sides demonstrated that the intestine model was able to absorb the APAP. There was a progressive APAP concentration decrease in the apical side (Figure 5A) concomitantly to APAP concentration increase at the intestinal basolateral side (Figure 5B). The maximum concentrations (Cmax) in the medium was around 2 µM for both static and dynamic conditions, after 12 h of the administration (Tmax).
For metabolism studies, the intravenous administration was mimicked by the application of 2 µM APAP in the medium of the liver compartment. The APAP concentration kinetics in the media under both static and dynamic conditions indicated that only at the dynamic conditions the decreases in the APAP concentration could be detected, reaching 0.87 µM APAP 12 h after 2 µM APAP administration (T1/2 = 12 h). The liver equivalents showed minimal metabolic efficiency under static conditions (Figure 5C). The APAP concentration reached 1.7 µM 12 h after APAP administration. The integrated, systemic like APAP absorption and metabolism evaluation was performed in the Intestine/Liver 2-OC model. The APAP was administered over the apical side of the intestine equivalent, emulating the oral route. Medium samples were collected from both Intestinal sides and also from the liver compartment. Figure 5D shows the progressive decay of the APAP concentration at the apical side in both static and dynamic conditions.
Figure 5E shows distinguishable absorption and metabolism phases. The flow also impacted in the intestinal absorption. The APAP Cmax in the medium changed from 2 µM on the “Intestine 2-OC” (Figure 5B) assembly to 1.7 µM for the dynamic “Intestine/Liver 2-OC” (Figure 5E). Figure 5F shows a direct comparison between the concentration–time profile of APAP in our dynamic “Intestine/Liver 2-OC” microphysiological system (red curve and y axis) and a representative profile obtained in humans after a single oral dose of 1000 mg (black curve and y axis).
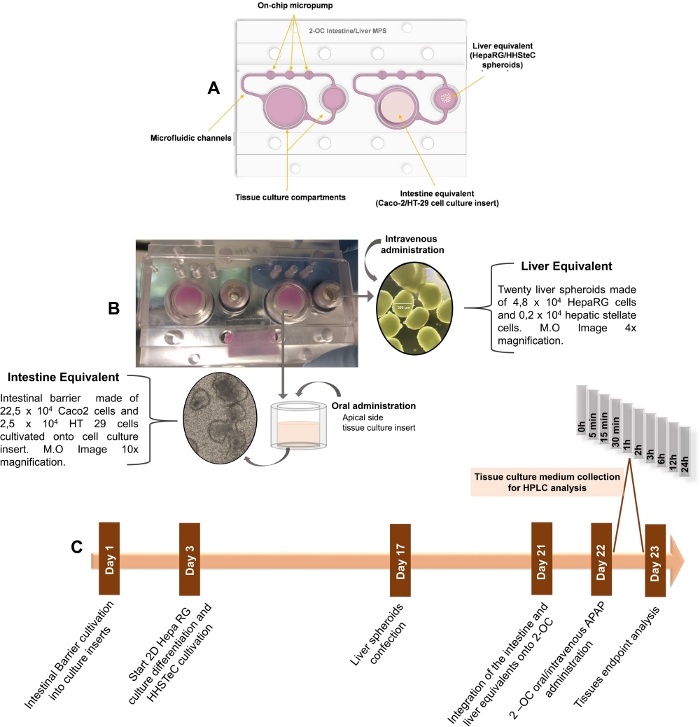
Figure 1: Schematic step compilation for PK studies in the 2-OC MPS. A) Schematic drawing of the 2-OC MPS, showing the intestinal and hepatic human tissue equivalents in bottom-up view. B) 2-OC MPS photograph with the intestinal and liver equivalents integrated into the device in bottom-up view, with representative optical microscopy images. C) Timeline of tissue equivalents preparation, APAP treatment, and culture medium collections phases for organoid manufacturing, and pharmacokinetic and toxicological assessments. This figure has been modified from Marin et al. Acetaminophen absorption and metabolism in an intestine/liver microphysiological system. Chem Biol Interact. 299, 59-76 (2019). Please click here to view a larger version of this figure.
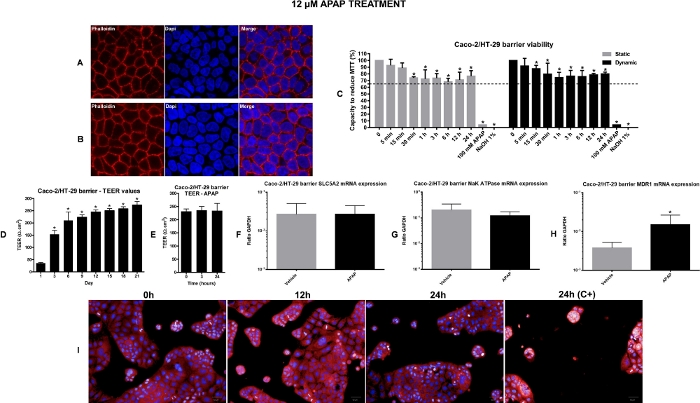
Figure 2: Viability and toxicological assessment of the intestine equivalents. A) Representative confocal fluorescence microscopy images of non-treated Caco-2/HT-29 cells stained with cell nuclei and actin filaments fluorescent dye (DAPI and Phalloidin respectively); 63x Magnification, zoom 2.6. B) Representative confocal fluorescence microscopy images of Caco-2/HT-29 cells treated with 12 µM APAP for 24 h, stained with nuclei and actin filaments fluorescent dye (DAPI and Phalloidin respectively); 63x Magnification, zoom 2.6. C) Intestine equivalents viability evaluation by MTT assay in both static and dynamic conditions. The values represented by the bars in the graph are percent calculated relative to vehicle control (time-point named as 0)*P<0.05 0 vs treatment. D) TEER evolution during the 21 days of differentiation. *P<0.05 day 1 vs other days. E) TEER values after APAP administration into the Intestine 2-OC MPS under dynamic conditions. Gene expression in intestines equivalents. Absorption potential of the intestine barrier and possible effects of 12 µM APAP for 24 h under dynamic condition was verified by SLC5A1 (F), Na-K-ATPase (G) and MDR1 (H) expression. Values represent the mean ± SEM of three independent experiments. The result is expressed as a ratio to housekeeping GAPDH. *P<0.05 vehicle vs APAP. I) Operetta image-based HCA performed by the Columbus® 2.4.0. software. Representative images of 2D intestine co-culture in different time points after 12 µM APAP treatment. Negative controls were medium (0 h) or vehicle (0.5% ethanol). Positive controls shown here is the 100 mM APAP. This figure has been modified from Marin et al. Acetaminophen absorption and metabolism in an intestine/liver microphysiological system. Chem Biol Interact. 299, 59-76 (2019). Please click here to view a larger version of this figure.
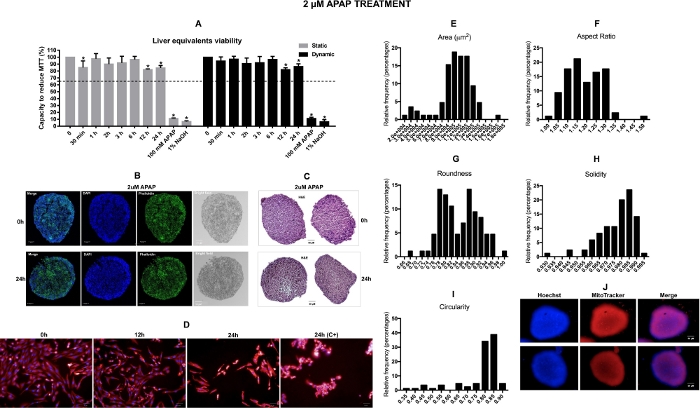
Figure 3: Viability and toxicological assessment of the liver equivalents. A) Liver equivalents viability evaluation by MTT assay in both static and dynamic conditions. The values represented by the bars in the graph are percent calculated relative to vehicle control (time-point named as 0) *P<0.05 0 vs treatment. B) Representatives confocal images captured from the vehicle and 2 µM APAP 24 h treated liver spheroids from an inner section. C) Representatives H&E (hematoxylin and eosin staining) images captured from the vehicle and 2 µM APAP 24 h treated liver spheroids from an inner section. Scale bar = 50 µm. D) Representative images of 2D liver co-culture in different time points after 2 µM APAP treatment. Samples treated with vehicle and with 2 µM APAP were considered in these analyses. The fluorophore dye mixture includes Hoechst for nuclear staining and Mitotracker Deep Red for mitochondria mass staining. Negative controls were medium (0 h) or vehicle (0.5% ethanol). Positive controls were 100 mM APAP. Measurements of whole spheroids images captured were performed using Fiji software. E) Frequency distributions of the area F) aspect ratio, G) roundness, H) solidity, I) circularity. N = 85. *p < 0,05. J) Representative images of 3D liver spheroids acquired by the Operetta using the LWD 10x objective. This figure has been modified from Marin et al. Acetaminophen absorption and metabolism in an intestine/liver microphysiological system. Chem Biol Interact. 299, 59-76 (2019). Please click here to view a larger version of this figure.
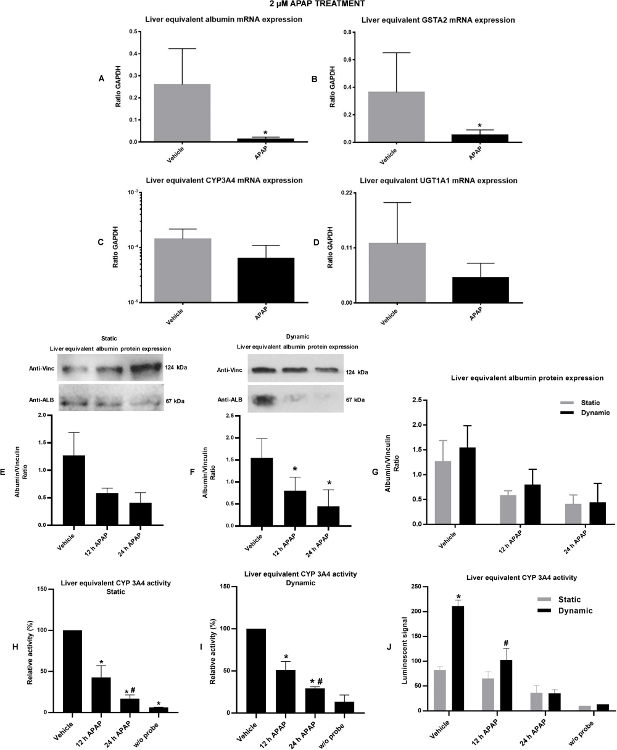
Figure 4: Liver viability/functionality and possible effects of 2 µM APAP under static and dynamic conditions over it were verified by gene and protein expression and by enzymatic activity. A) albumin gene expression. B) GSTA2 gene expression. Liver capability to perform phase I and phase II metabolism and possible effects of 2 µM APAP for 24 h under dynamic condition over it were verified by gene expression of CYP3A4 (C) and by UGT1A1 (D) respectively. E) Total albumin protein expression under static condition. F) Total albumin protein expression under dynamic conditions. condition. G) Comparative graph illustrating the difference in total albumin expression in liver equivalents cultivated and treated under static or dynamic conditions. H) CYP 3A4 in vitro enzymatic activity under static conditions. I) CYP 3A4 in vitro enzymatic activity under dynamic conditions. J) Comparative graph illustrating the difference in CYP 3A4 activity in liver equivalents cultivated and treated under static or dynamic conditions. Values represent the mean ± SEM of three independent experiments. The data of gene expression is expressed as a ratio to housekeeping GAPDH. The data of protein expression is expressed as a ratio to vinculin protein. *P<0.05 vehicle vs APAP treatment. This figure has been modified from Marin et al. Acetaminophen absorption and metabolism in an intestine/liver microphysiological system. Chem Biol Interact. 299, 59-76 (2019). Please click here to view a larger version of this figure.
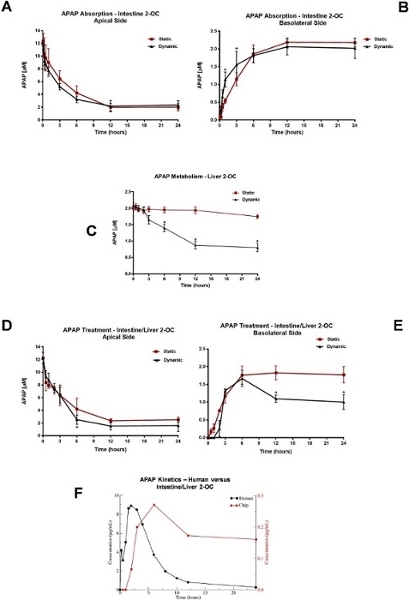
Figure 5: Analyzes of APAP pharmacokinetics in 2-OC MPS. APAP absorption profile after 12 µM APAP administration at the apical side of the Intestine 2-OC MPS preparation. The intestinal barrier was made of a coculture of Caco-2/HT-29 cell lines (A) Static and dynamic APAP concentrations in the medium from the apical side (representing the human intestinal lumen side). (B) Static and dynamic APAP concentrations in the medium from the basolateral side (representing the human intestinal bloodstream side). *P<0.05 static vs dynamic conditions. C) APAP metabolism profile in the Liver 2-OC MPS by HepaRG/HHSTeC liver spheroids. Comparison of static and dynamic conditions after a 2 µM APAP administration into the medium. *P<0.05 0h vs 6 h, 12 h, and 24 h APAP treatment. APAP absorption and metabolism profile after 12 µM administration on the intestinal barrier apical side of the Intestine/Liver 2-OC MPS preparation. This emulates the oral route. The intestinal barrier was made of Caco-2/HT-29 cell lines and the liver equivalent made of spheroids of HepaRG/HHSTeC cell lines. (D) Intestinal/Liver 2-OC APAP concentrations in the apical side of the intestinal barrier under static and dynamic conditions. (E) Intestinal/Liver 2-OC APAP concentrations in the medium under static and dynamic conditions. *P<0.05 static vs dynamic conditions. (F) Comparison between the concentration–time profile of APAP in our microphysiological system (red curve and y axis) and a representative profile obtained in humans after a single oral dose of 1000 mg (black curve and y axis). Data was extracted from plots using WebPlotDigitizer 4.2 (https://automeris.io/WebPlotDigitizer). This figure has been modified from Marin et al. Acetaminophen absorption and metabolism in an intestine/liver microphysiological system. Chem Biol Interact. 299, 59-76 (2019). Please click here to view a larger version of this figure.
| HPLC system | Waters Alliance 2695 (Milford, MA, USA), equipped with quaternary pump, sample manager and degasser | ||
| Detector | Waters 2996 Uv-Vis set in 210-400 nm range | ||
| System control, data acquisition, and processing | Waters Empower 2002 chromatography software | ||
| Column | Reversed-phase Luna C18 | ||
| (150 x 4.6 mm I.D.; 5mm particle size) | |||
| Phenomenex | |||
| Guard Column | Reversed-phase Luna C18 (4 x 3 mm) | ||
| Phenomenex | |||
| Mobile phase | Solvent A- Acetonitrile | ||
| Solvent B- 0.10 M ammonium acetate, pH 6.8 | |||
| Isocratic conditions | Time | A (%) | B (%) |
| (min.) | |||
| 15 | 5 | 95 | |
| Flow | 1.0 mL/min | ||
| Injection volume | 25 μL | ||
| Temperature | 25 °C | ||
| APAP Detection | UV @ 243 and 254 nm | ||
| Run time | 15 minutes | ||
Table 1: Conditions and parameters to be used for HPLC-UV analyses of APAP in culture medium matrices.
| Nominal concentration | Calculated APAP concentration (µM) | Average (µM) | S.D.b | C.V. | DEV | |||||
| (µM) | (Triplicate of each concentration) | (µM) | (%)c | (%)d | ||||||
| Assay number | 1 | 2 | 3 | 4 | 5 | 6 | ||||
| 0.25 (LOD) | 0.02 | 0.08 | 0.21 | 0.14 | 0.08 | 0.27 | 0.13 | 0.11 | 84.96 | + 46.05 |
| 0.50 (LLOQ) | 0.31 | 0.36 | 0.29 | 0.47 | 0.42 | 0.65 | 0.41 | 0.05 | 12.72 | + 17.08 |
| 1.00 | 0.87 | 0.87 | 0.80 | 1.04 | 1.02 | 1.01 | 0.93 | 0.04 | 3.76 | + 6.65 |
| 2.50 | 2.44 | 2.61 | 2.52 | 2.42 | 2.56 | 2.54 | 2.52 | 0.04 | 1.54 | -0.62 |
| 5.00 | 5.02 | 4.99 | 5.06 | 5.01 | 5.01 | 5.05 | 5.02 | 0.09 | 1.88 | -0.45 |
| 10.00 | 10.21 | 10.13 | 9.96 | 10.25 | 10.08 | 10.21 | 10.14 | 0.10 | 0.97 | -1.41 |
| 25.00 | 25.33 | 25.28 | 25.20 | 25.13 | 25.14 | 24.92 | 25.17 | 0.36 | 1.45 | -0.67 |
| 50.00 | 50.30 | 50.04 | 50.51 | 49.70 | 49.98 | 49.19 | 49.95 | 0.86 | 1.71 | +0.09 |
| 100.00 | 99.75 | 99.90 | 99.70 | 100.09 | 99.97 | 100.40 | 99.97 | 0.69 | 0.69 | +0.03 |
| R2 | 1.00 | 1.00 | 0.9999 | 1.00 | 1.00 | 0.9999 | ||||
Table 2: Inter-run variation – accuracy, precision, and linearity of standard curve samples prepared in a mixture of DMEM medium with 0.10 M ammonium acetate buffer (1:1, v/v) from six separate assays.a
aA linear curve was fitted to the data for a response (APAP) versus theoretical concentration as described in Experimental. The calculated concentration was derived from reading the response for each standard sample against the calibration curve. Each entry (assay 1-6) corresponds to the average value of triplicate analysis.
bSD= Standard deviation.
cC.V. (coefficient of variation. precision).
dAccuracy (DEV %) = the deviation of the calculated concentration from the nominal value.
This figure has been modified from Marin et al. Acetaminophen absorption and metabolism in an intestine/liver microphysiological system. Chem Biol Interact. 299, 59-76 (2019).
| Nominal concentration | Calculated APAP concentration (µM) | Average | S.D.b | C.V. | DEV | ||||
| (µM) | (Triplicate of each concentration) | (µM) | (µM) | (%)c | (%)d | ||||
| Assay number | 1 | 2 | 3 | 4 | 5 | ||||
| 0.25 (LOD) | 0.34 | 0.12 | 0.30 | 0.16 | 0.00 | 0.18 | 0.08 | 45.46 | +26.03 |
| 0.50 (LLOQ) | 0.36 | 0.44 | 0.49 | 0.43 | 0.40 | 0.43 | 0.02 | 5.84 | +14.97 |
| 1.00 | 0.87 | 0.98 | 1.04 | 0.94 | 0.83 | 0.93 | 0.04 | 4.67 | +6.85 |
| 2.50 | 2.41 | 2.46 | 2.49 | 2.52 | 2.43 | 2.46 | 0.06 | 2.39 | +1.56 |
| 5.00 | 5.00 | 4.99 | 5.12 | 5.10 | 5.14 | 5.07 | 0.15 | 3.00 | -1.38 |
| 10.00 | 10.08 | 10.01 | 9.93 | 10.10 | 10.29 | 10.08 | 0.18 | 1.80 | -0.81 |
| 25.00 | 25.14 | 25.18 | 24.96 | 25.32 | 25.35 | 25.19 | 0.45 | 1.78 | -0.76 |
| 50.00 | 50.19 | 50.23 | 49.83 | 49.56 | 50.17 | 49.99 | 0.87 | 1.75 | +0.01 |
| 100.00 | 99.87 | 99.84 | 100.10 | 100.13 | 99.80 | 99.95 | 2.12 | 2.12 | +0.05 |
| R2 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
Table 3: Inter-run variation – accuracy, precision, and linearity of standard curve samples prepared in a mixture of Williams medium with 0.10 M ammonium acetate buffer (1:1, v/v) from six separate assays.a
aA linear curve was fitted to the data for a response (APAP) versus theoretical concentration as described in Experimental. The calculated concentration was derived from reading the response for each standard sample against a calibration curve. Each entry (assay 1-5) corresponds to the average value of triplicate analysis.
bSD= Standard deviation.
cC.V. (coefficient of variation. precision).
dAccuracy (DEV %) = the deviation of the calculated concentration from the nominal value.
This figure has been modified from Marin et al. Acetaminophen absorption and metabolism in an intestine/liver microphysiological system. Chem Biol Interact. 299, 59-76 (2019).
| Williams medium | Nominal concentration | Measured concentration | S.D. | C.V. | DEV |
| (µM) | (µM) | (µM) | (%) | (%) | |
| Intra-run (n=3) | 0.50 (LLOQ) | 0.49 | 0.08 | 15.55 | +1.77 |
| 4.50 | 4.59 | 0.23 | 5.10 | -2.06 | |
| 45.00 | 41.23 | 0.76 | 1.85 | +8.37 | |
| 90.00 | 82.29 | 1.75 | 2.13 | +8.57 | |
| Inter-run (n=15) | 0.50 (LLOQ) | 0.43 | 0.05 | 10.99 | +14.97 |
| 4.50 | 4.37 | 0.19 | 4.42 | +2.99 | |
| 45.00 | 42.35 | 0.82 | 1.93 | +5.88 | |
| 90.00 | 85.22 | 2.25 | 2.65 | +5.31 | |
| DMEM medium | Nominal concentration | Measured concentration | S.D. | C.V. | DEV |
| (µM) | (µM) | (µM) | (%) | (%) | |
| Intra-run (n=3) | 0.50 (LLOQ) | 0.47 | 0.09 | 18.77 | +6.35 |
| 4.50 | 4.45 | 0.30 | 6.63 | +1.04 | |
| 45.00 | 44.24 | 1.59 | 3.58 | +1.69 | |
| 90.00 | 86.40 | 4.09 | 4.73 | +4.00 | |
| Inter-run (n=12) | 0.50 (LLOQ) | 0.46 | 0.08 | 17.81 | +7.75 |
| 4.50 | 5.16 | 0.27 | 5.31 | -14.68 | |
| 45.00 | 48.99 | 2.10 | 4.29 | -8.86 | |
| 90.00 | 96.18 | 4.47 | 4.65 | -6.86 | |
Table 4: Intra- and inter-run precision and accuracy for APAP in quality control samples.a
aThe data are shown as averages. SD (standard deviation). C.V. (coefficient of variation. precision) and accuracy (percent deviation. DEV%).
This figure has been modified from Marin et al. Acetaminophen absorption and metabolism in an intestine/liver microphysiological system. Chem Biol Interact. 299, 59-76 (2019).
| Primer (5’ → 3’) | |||||
| Tissue | Gene | Forward | Reverse | ||
| Intestine | SGLT1/SLC5A1 | gAgCCCAgCAACTgTCCCAC | CAggCTCCAACACAgACggT | ||
| NA-K-ATPase | ACCgCCCAgAAATCCCAAAAC | CAgCggTCATCCCAgTCC | |||
| MDR1 | TggATgTTTCCggTTTggAg | TgTgggCTgCTgATATTTTgg | |||
| Liver | Albumin | TgCAAggCTgATAAggAg | TTTAgACAgggTgTTggCTTTACAC | ||
| GSTA2 | CTgAggAACAAgATgCCAAgC | AgCAgAgggAAggCTggAAATAAg | |||
| CPY3A4 | ggAAggTggACCCAgAAACTgC | TTACggTgCCATCCCTTgAC | |||
| UGT1A1 | ATgCAAAgCgCATggAgAC | ggTCCTTgTgAAggCTggAg | |||
Table 5: Real-time qPCR primers to evaluate gene transcription at mRNA level in the 2-OC cultures for intestine and liver tissues
Discussion
The accurate and reliable assessment of the pharmacologic properties of investigative new drugs is critical for reducing the risk in the following development steps. The MPS is a relatively new technology, that aims at improving the predictive power and reducing the costs and time spent with preclinical tests. Our group is advancing in the assessment of pharmacokinetic and toxicological properties mostly needed for lead optimization. We worked with the 2-OC microfluidic device, which has two chambers, allowing the integration of two organoids. APAP was chosen because it has plenty of high-quality human data, is mostly metabolized by the liver, and also displays hepatotoxic properties. The study protocol aimed to emulate some steps of the human phase I clinical trial, where a drug is administered orally to volunteers, and periodic blood samples are drawn to assess the drugs’ concentration to know the pharmacokinetic properties and biochemical and clinical parameters are collected to assess the safety and tolerability. Therefore, when the MPS evolves enough to provide reliable predictions, it will significantly reduce the risk of failure in the phase I trial. By adding the disease models in the future, the same assumption possibly will apply to the phases II and III clinical trials.
All studies were performed in three models: Intestine 2-OC (APAP oral administration), Liver 2-OC (Intravenous administration), and Intestine/Liver 2-OC (oral administration). The first model isolated the absorption, the second the metabolism, and the third integrated both. We first produced the organoids and incorporated into the microfluidic device, second administered the APAP and collected the media samples, and last performed a set of tests to assess the cell viability and functionality and the toxicological impact of APAP exposure. For the pharmacokinetic studies, we developed and validated a chromatographic method for APAP quantification in the medium. The validation complied with the Guideline on Bioanalytical Method Validation regarding specificity, linearity, the limit of quantification (LOQ), the limit of detection (LOD), matrix effect, precision, and accuracy, as outlined by FDA29,30. The specificity was established with blank, pooled, and individual biological samples from two different sources. The method performed acceptably during the analysis, and the data confirmed the ability of a simple isocratic mobile phase to separate and quantify APAP.
The viability/functionally of the intestine and liver organoids was assessed by different techniques. For the intestine equivalent, the confocal fluorescence microscopy (Figure 2A-B), the MTT assay (Figure 2C), gene expression assessment (Figure 2D-F) or HCA experiment, did not detect any toxicity 24 h after APAP exposure. Likewise, MTT assay did not detect toxic insults induced by APAP in liver equivalents (Figure 3A). However, the HCA technique added the possibility of detection of very early toxic events (24 h after APAP exposure) for liver cells, by the observation of cell phenotypic changes, with some mechanistic clues (Figure 3D). On the other hand, the confocal fluorescence microscopy (Figure 3B) and histology (Figure 3C) were showed to be a useful complementary tool in the investigation of the viability status of the human tissue’s equivalents. For the liver cells, the simultaneous use of different techniques was very advantageous. The MTT assay, as mentioned before, was not able to detect the APAP cytotoxicity detected by the HCA 24 h after APAP exposure. The gene expression analyses assessed the functionality of the intestine (Figure 2D-F) and liver organoids (Figure 4A-D) at both basal and 24 h after APAP treatment. Under basal conditions, there were normal levels of intestinal and liver-specific markers, suggesting proper functionality. After 24 h of APAP exposure, there was a downregulation of albumin, GST mRNA levels, and a tendency to reduce the CYP3A4 gene expression in liver equivalent tissues, indicating APAP early cytotoxicity. Corroborating these findings, Western blotting experiments showed that the reduction in gene expression was also accompanied by a robust reduction in liver total albumin protein expression in response to APAP treatment (Figure 4E-F), confirming the toxic insults imposed on liver tissue by exposure to APAP. Accordingly, experiments of in vitro enzymatic activity showed a robust and progressive reduction in CYP 3A4 activity levels induced by APAP treatment, in liver equivalents (Figure 4H-I).
Morphometrical statistics of organoids were performed on Image J (Figure 3E-I). The area reveals how close the 2D size is to all organoids analyzed, which could be used as standardization in this protocol so that an unbiased result can be produced. The ‘shape descriptors’ reveal statistics corresponding precisely to the shape morphology. The Aspect Ratio is an index which uses the major and minor axis, so results around 1 indicate no bias (i.e., preferential growth) during organoid formation. Values of Roundness (4 ×[area]/ (π × [major axis]2)) are very sensitive to a preferential growth, which would be revealed as a major axis. Solidity ([area]/ ([convex area])) is essential in showing gross morphology as it is not affected by irregularities in borders since it uses convex area (=envelope). Distributions centered around 1.0 indicate putative spherical growth. Conversely, Circularity (4× π [area]/[perimeter]2) is very sensitive to a complex perimeter, so “cavities” or “pockets” would impact this index. Thus, circularity around 1 also corroborates putative spherical growth, compatible with proper organoid functionality.
The analytical results showed that the MPS could emulate the APAP absorption and metabolism properties, both isolated or integrated in a curve comparable to that produced in vivo (Figure 5F) without the excretion phase. The APAP absorption was similar after oral administration to both Intestine MPS or Intestine/Liver MPS models, under both static and dynamic conditions (Figure 5A-B, Figure 5D-E). In both, there was an APAP concentration decreases at apical side concomitantly to its increase at the basolateral side, with no significant difference for static and dynamic conditions. In contrast, the APAP hepatic metabolism differed under these conditions. The circulating media in MPS seemed to improve the organoid metabolic capability (Figure 5C and Figure 5E). There was a significant APAP decay underflow not seen without flow. Interestingly, in vitro, CYP3A4 activity experiments corroborate the hypothesis that the presence of flow in the system increases the functionality of human liver equivalents. As shown in the graph in Figure 4J, the activity of CYP 3A4 was significantly higher in liver equivalents maintained under flow at both basal and APAP treatment conditions. Likewise, the liver equivalents kept under flow (dynamic) showed a tendency to increase the protein expression of albumin when compared to those kept without flow (static) both at baseline or at APAP treated conditions (Figure 4G).
When compared to the concentration-time profile after oral administration of APAP to humans, our microphysiological system shows much larger t1/2 (half-life time) (Figure 5F). This happens, as mentioned before, because while we have a two-organ system with intestine and liver equivalents that can absorb and metabolize the drug, there is no kidney equivalent to excrete APAP and its metabolites from the plasma compartment38. In addition to that, the microphysiological system shows smaller Cmax (peak plasma concentration) and larger tmax (time to reach Cmax) than what is typically observed for humans (Figure 5F). This is a consequence of the differences in the scale of the in vitro and in vivo experiments. Overall, there are three main differences between the concentration–time profile obtained using the microphysiological system and profiles obtained after oral administration of APAP to humans: larger t1/2, smaller Cmax and larger tmax (Figure 5F). While the larger t1/2 is due to the absence of a kidney equivalent to excrete APAP and its metabolites from the plasma equivalent, smaller Cmax and larger tmax are consequences of the small scale of the experiment when compared to the human body. The best scaling strategies for building and operating microphysiological systems are still an active area of research and it is unlikely that a single approach is optimal for all systems39. Additionally, mathematical modeling or machine learning can be used to apply corrections or learn the mapping from the micro scale to full scale in order to extrapolate in vitro data obtained with the microphysiological systems to the in vivo behavior observed in humans40.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thanks to Dr. Christie Guguen-Guillouzo, Dr. Philippe Gripon at Unit 522 INSERM and to Dr. Christian Trepo at Unit 271 INSERM for the use of the Biological Material (Hepa RG cells) and for making then available for us in order to perform the academic research.
Materials
| 1x DPBS | Thermo Fisher Scientific | 14190235 | No calcium, no magnesium |
| 2-OC | TissUse GmbH | Two-organ chip | |
| 384-well Spheroid Microplate | Corning | 3830 | Black/Clear, Round Bottom, Ultra-Low Attachment |
| 4% Paraformaldehyde | Use to fix cell | ||
| Acetaminophen | Sigma Aldrich | A7085 | Use to MPS assays |
| Acetonitrile | Tedia | Used to perform HPLC | |
| Alexa Fluor 647 phalloidin | Thermo Fisher Scientific | confocal experiment | |
| Ammonium acetate | Sigma Aldrich | Used to perform HPLC | |
| Caco-2 cells | Sigma Aldrich | 86010202 | |
| Cacodylate buffer | |||
| Cell culture flasks | Sarstedt | ||
| Confocal Fluorescence microscope | Leica | DMI6000 | |
| Cryostat | Leica | CM1950 | |
| DMEM high glucose | Thermo Fisher Scientific | 12800017 | Add supplements: 10% fetal bovine serum, 100 units per mL penicillin, 100 µg/mL streptomycin, and 1% non-essential amino acids |
| DMSO | Sigma Aldrich | D4540 | Add 2% to HepaRG media |
| Ethanol | Synth | ||
| Fetal Bovine Serum | Thermo Fisher Scientific | 12657029 | |
| Freezing medium OCT | Tissue-Tek | Tissue-Tek® O.C.T.™ Compound is a formulation of watersoluble glycols and resins, providing a convenient specimen matrix for cryostat sectioning at temperatures of -10°C and below. | |
| Hematoxylin & Eosin | |||
| HepaRG cells | Biopredic International | HPR101 | Undifferentiated cells |
| HHSTeC | ScienCell Research Laboratories | 5300 | Cells and all culture supplements |
| Hoechst 33342 | HCA experiments | ||
| HT-29 cells | Sigma Aldrich | 85061109 | |
| Human Insulin | Invitrogen – Thermo Fisher Scientific | 12585014 | |
| Hydrocortisone | Sigma Aldrich | H0888 | |
| Isopropanol | Merck | 278475 | |
| Karnovsky’s fixative | |||
| L-glutamine | Thermo Fisher Scientific | A2916801 | |
| Luna C18 guard column SS | Phenomenex | Used to perform HPLC | |
| Microscope | Leica | DMi4000 | |
| Microtome | Leica | RM2245 | |
| Millicell 0.4 µm pore size inserts | Merck | PIHP01250 | |
| Millicell ERS-2 meter | Merck | MERS00002 | Used to TEER measurement |
| MitoTracker Deep Red | HCA experiments | ||
| MTT | Thermo Fisher Scientific | M6494 | |
| MX3000P system | Agilent Technologies | ||
| Neubauer chamber | Counting cells | ||
| Operetta High Content Imaging System | Perkin Elmer | Used to perform HCA | |
| P450-Glo CYP3A4 Assay with Luciferin-IPA | Promega | Cat.# V9001 | |
| Penicillin/Streptomycin | Thermo Fisher Scientific | 15070063 | Cell culture |
| Permount | Thermo Fisher Scientific | Histology | |
| Primers | RT-qPCR | ||
| PVDF membrane | BioRad | ||
| PVDF Syringe filter | 0.22 μm pore size | ||
| Reversed-phase Luna C18 column | Phenomenex | Used to perform HPLC | |
| Shaker (IKA VXR Basic Vibrax) | IKA Works GmbH & Co | 2819000 | Used for spheroids to improve MTT assay |
| Stellate Cell Media (STeC CM) | ScienCell | 5301 | Add STeC CM supplements |
| SuperScriptIITM Reverse Transcriptase | Thermo Fisher Scientific | ||
| SYBR Green PCR Master Mix | Thermo Fisher Scientific | ||
| TRizol TM reagent | Thermo Fisher Scientific | Trizol is a monophasic solution of phenol and guanidine isothiocyanate. | |
| Trypsin/EDTA solution | Thermo Fisher Scientific | R001100 | |
| Ultra-low-attachment plates | Corning | CLS3471-24EA | 6 wells |
| Vectashield plus DAPI mounting media | |||
| White Opaque 96-well Microplate | PerkinHelmer | ||
| Wide-bore tips | |||
| Williams E | Pan Biotech | P04-29510 | Add supplements: 10% fetal bovine serum, 2 mM L-glutamine, 100 units per ml penicillin, 100 µg/mL streptomycin and 5 µg/mL human insulin |
References
- Zhang, D., Luo, G., Ding, X., Lu, C. Preclinical experimental models of drug metabolism and disposition in drug discovery and development. Acta Pharmaceutica Sinica B. 2 (6), 549-561 (2012).
- Hynds, R. E., Giangreco, A. The relevance of human stem cell-derived organoid models for epithelial translational medicine. Stem Cells. 31 (3), 417-422 (2013).
- Sivaraman, A., et al. A Microscale In vitro Physiological Model of the Liver: Predictive Screens for Drug Metabolism and Enzyme Induction. Current Drug Metabolism. 6 (6), 569-591 (2005).
- Dehne, E. M., Hasenberg, T., Marx, U. The ascendance of microphysiological systems to solve the drug testing dilemma. Future Science OA. 3 (2), 185 (2017).
- Oleaga, C., et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Scientific Reports. 6, 20030 (2016).
- Maschmeyer, I., et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 15 (12), 2688-2699 (2015).
- Esch, M. B., Mahler, G. J., Stokol, T., Shuler, M. L. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip. 14 (16), 3081-3092 (2014).
- Materne, E. M., et al. The Multi-organ Chip – A Microfluidic Platform for Long-term Multi-tissue Coculture. Journal of Visualized Experiments. , e52526 (2015).
- Esch, M. B., Ueno, H., Applegate, D. R., Shuler, M. L. Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab Chip. 16 (14), 2719-2729 (2016).
- Sung, J. H., Kam, C., Shuler, M. L. A microfluidic device for a pharmacokinetic–pharmacodynamic (PK-PD) model on a chip. Lab on a Chip. 10 (4), 446-455 (2010).
- Viravaidya, K., Sin, A., Shuler, M. L. Development of a Microscale Cell Culture Analog to Probe Naphthalene Toxicity. Biotechnology Progress. 20 (1), 316-323 (2004).
- Sin, A., Chin, K. C., Jamil, M. F., Kostov, Y., Rao, G., Shuler, M. L. The Design and Fabrication of Three-Chamber Microscale Cell Culture Analog Devices with Integrated Dissolved Oxygen Sensors. Biotechnology Progress. 20 (1), 338-345 (2004).
- Tsamandouras, N., et al. Integrated Gut and Liver Microphysiological Systems for Quantitative In vitro Pharmacokinetic Studies. The AAPS Journal. 19 (5), 1499-1512 (2017).
- Graham, G. G., Davies, M. J., Day, R. O., Mohamudally, A., Scott, K. F. The modern pharmacology of paracetamol: Therapeutic actions, mechanism of action, metabolism, toxicity and recent pharmacological findings. Inflammopharmacology. 21 (3), 201-232 (2013).
- Hodgman, M. J., Garrard, A. R. A Review of Acetaminophen Poisoning. Critical Care Clinics. 28 (4), 499-516 (2012).
- Maschmeyer, I., et al. Chip-based human liver-intestine and liver-skin co-cultures – A first step toward systemic repeated dose substance testing in vitro. European Journal of Pharmaceutics and Biopharmaceutics. 95, 77-87 (2015).
- Bessems, J. G. M., Vermeulen, N. P. E. Paracetamol (acetaminophen)-induced toxicity: Molecular and biochemical mechanisms, analogues and protective approaches. Critical Reviews in Toxicology. 31 (1), 55-138 (2001).
- Hirt, M. N., et al. Increased afterload induces pathological cardiac hypertrophy: A new in vitro model. Basic Research in Cardiology. 107 (6), 307 (2012).
- Fink, C., et al. Chronic stretch of engineered heart tissue induces hypertrophy and functional improvement. The FASEB journal official publication of the Federation of American Societies for Experimental Biology. 14 (5), 669-679 (2000).
- Zhang, B., Peticone, C., Murthy, S. K., Radisic, M. A standalone perfusion platform for drug testing and target validation in micro-vessel networks. Biomicrofluidics. 7 (4), 044125 (2013).
- Araújo, F., Sarmento, B. Towards the characterization of an in vitro triple co-culture intestine cell model for permeability studies. International Journal of Pharmaceutics. 458 (1), 128-134 (2013).
- Cadena-Herrera, D., et al. Validation of three viable-cell counting methods: Manual, semi-automated, and automated. Biotechnology Reports. 7, 9-16 (2015).
- Gripon, P., et al. Infection of a human hepatoma cell line by hepatitis B virus. Proceedings of the National Academy of Sciences of the United States of America. 99 (24), 15655-15660 (2002).
- Guillouzo, A., et al. The human hepatoma HepaRG cells: A highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chemico-Biological Interactions. 168 (1), 66-73 (2007).
- Wagner, I., et al. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab on a Chip. 13 (18), 3538-3547 (2013).
- Forrest, J. A. H., Clements, J. A., Prescott, L. F. Clinical Pharmacokinetics of Paracetamol. Clinical Pharmacokinetics. 7 (2), 93-107 (1982).
- Dollery, C. . Therapeutic Drugs. , (1999).
- Shah, V. P., et al. Bioanalytical method validation-a revisit with a decade of progress. Pharmaceutical research. 17 (12), 1551-1557 (2000).
- Marin, T. M., et al. Shp2 negatively regulates growth in cardiomyocytes by controlling focal adhesion kinase/src and mTOR pathways. Circulation Research. 103 (8), 813-824 (2008).
- Linkert, M., et al. Metadata matters: Access to image data in the real world. Journal of Cell Biology. 189 (5), 777-782 (2010).
- OECD. OECD TG 491 – Short Time Exposure In vitro Test Method for Identifying i) Chemicals Inducing Serious Eye Damage and ii) Chemicals Not Requiring Classification for Eye Irritation or Serious Eye Damage. OECD. 14, (2018).
- Fernandes, M. B., Gonçalves, J. E., Tavares, L. C., Storpirtis, S. Caco-2 cells permeability evaluation of nifuroxazide derivatives with potential activity against methicillin-resistant Staphylococcus aureus (MRSA). Drug Development and Industrial Pharmacy. 41 (7), 1066-1072 (2015).
- OECD. OECD TG 492 – Reconstructed human Cornea-like Epithelium (RhCE) test method for identifying chemicals not requiring classification and labelling for eye irritation or serious eye damage. OECD. 35, (2018).
- OECD, O. E. C. D. OECD TG 431 – In vitro skin corrosion: reconstructed human epidermis (RHE) test method. OECD. 8, (2016).
- OECD. OECD TG 439 – In vitro Skin Irritation: Reconstructed Human Epidermis Test Method. OECD. 21, (2015).
- Marin, T. M., et al. Acetaminophen absorption and metabolism in an intestine/liver microphysiological system. Chemico-Biological Interactions. 299, 59-76 (2019).
- Maass, C., Stokes, C. L., Griffith, L. G., Cirit, M. Multi-functional scaling methodology for translational pharmacokinetic and pharmacodynamic applications using integrated microphysiological systems (MPS). Integrative Biology (United Kingdom). 9 (4), 290-302 (2017).
- Sung, J. H., Wang, Y., Shuler, M. L. Strategies for using mathematical modeling approaches to design and interpret multi-organ microphysiological systems (MPS). APL Bioengineering. 3 (2), 021501 (2019).

