T and B Cell Receptor Immune Repertoire Analysis using Next-generation Sequencing
Summary
The current protocol describes a method for DNA isolation from blood samples and intestinal biopsies, generation of TCRβ and IGH PCR libraries for next-generation sequencing, performance of a NGS run and basic data analysis.
Abstract
Immunological memory, the hallmark of adaptive immunity, is orchestrated by T and B lymphocytes. In circulation and different organs, there are billions of unique T and B cell clones, and each one can bind a specific antigen, leading to proliferation, differentiation and/or cytokine secretion. The vast heterogeneity in T and B cells is generated by random recombination of different genetic segments. Next-generation sequencing (NGS) technologies, developed in the last decade, enable an unprecedented in-depth view of the T and B cell receptor immune repertoire. Studies in various inflammatory conditions, immunodeficiencies, infections and malignancies demonstrated marked changes in clonality, gene usage, and biophysical properties of immune repertoire, providing important insights about the role of adaptive immune responses in different disorders.
Here, we provide a detailed protocol for NGS of immune repertoire of T and B cells from blood and tissue. We present a pipeline starting from DNA isolation through library preparation, sequencing on NGS sequencer and ending with basic analyses. This method enables exploration of specific T and B cells at the nucleotide or amino-acid level, and thus can identify dynamic changes in lymphocyte populations and diversity parameters in different diseases. This technique is slowly entering clinical practice and has the potential for identification of novel biomarkers, risk stratification and precision medicine.
Introduction
The adaptive immune system, comprised of T and B lymphocytes, utilizes immunological memory to recognize a previously encountered antigen and initiate a rapid response. Lymphocytes are generated in the bone marrow and mature in the thymus (T cells) or bone marrow (B cells). Both the T cell receptor (TCR) and B cell receptor (BCR) display unique configurations that allow recognition of specific antigens. In homeostasis, T and B cells constantly circulate and survey the trillions of different peptides presented on antigen-presenting cells. TCR or BCR ligation of a specific antigen with high affinity, together with appropriate co-stimulation, leads to cell activation, resulting in cytokine secretion, clonal expansion and generation of antibodies, in the case of B cells.
The enormous array of the different T or B cells is collectively termed immune repertoire, enabling recognition of countless of different epitopes. In order to generate such a vast repertoire, a complex process of random assembly of different gene segments takes place, creating nearly endless combinations of receptors that can bind unique antigens1. This process, called V(D)J recombination, includes rearrangements of different variable (V), diversity (D) and joining (J) genes, accompanied by random deletions and insertions of nucleotides in the junctions2.
The architecture of the adaptive immune system has interested scientists in different fields for many decades. In the past, Sanger sequencing, complementary determining region 3 (CDR3) spectratyping, and flow cytometry were used to characterize the immune repertoire, but provided low resolution. In the last decade, advances in next-generation sequencing (NGS) methods enabled in-depth insight into the characteristics and composition of an individual’s TCR and BCR repertoires3,4. These high-throughput systems (HTS) sequence and process millions of rearranged TCR or BCR products simultaneously and permit a high-resolution analysis of specific T and B cells at the nucleotide or amino acid level. NGS provides a new strategy to study the immune repertoire in both health and disease. Studies utilizing HTS demonstrated altered TCR and BCR repertoires in autoimmune diseases5, primary immunodeficiencies6,7, and malignancies, such as in acute myeloid leukemia8. Using NGS, we and others have shown oligoclonal expansion of specific T and B cell clones, in patients with inflammatory bowel disease (IBD), including ulcerative colitis and Crohn’s disease9,10,11,12,13,14. Overall, studies from different fields suggest changes in the repertoire have a crucial role in the pathogenesis of immune-mediated disorders.
The current protocol describes a method for isolation of DNA from intestinal biopsies and blood, generation of TCRβ and IGH PCR libraries for NGS, and performance of sequencing run. We also provide basic steps in immune repertoire data analysis. This protocol can be applied for the generation of TCRα, TCRγ, and IGL libraries as well. The method is also compatible with other organs (e.g., lymph nodes, tumors, synovial fluid, fat tissue, etc.) as long as tissue-specific digestion protocols are used.
Protocol
This study was approved by the institutional review board at Sheba Medical Center, and informed written consent was obtained from all participating subjects.
1. DNA isolation and quantification
- Digestion and cell lysis of intestinal biopsies
- Retrieve intestinal biopsies, either freshly collected or those stored at -20 °C or -80 °C. If using frozen biopsies, thaw on ice.
- Add 600 µL of nuclei lysis solution to a sterile 1.7 mL micro-centrifuge tube, chilled on ice.
- Place biopsy into a microcentrifuge tube with lysis solution and incubate at 65 °C for 15-30 min.
- Add 17.5 µL of 20 mg/mL proteinase K and incubate overnight at 55 °C, with gentle shaking.
- Allow sample to cool to room temperature for 5 min before proceeding to step 1.3.
- Cell lysis of whole blood
- Obtain 3 mL of whole blood in EDTA-, heparin-, or citrate-containing tube.
- Gently rock tube until thoroughly mixed, and transfer blood to a sterile 15 mL centrifuge tube containing 9 mL of cell lysis solution. Invert several times during a 10 min incubation period at room temperature.
- Centrifuge at 2,000 x g for 10 min at room temperature, and then discard as much supernatant as possible without disturbing the visible white pellet. Approximately 50–100 μL of residual liquid will remain.
- Vortex tube vigorously for 15 s until completely resuspended. Add 3 mL of nuclei lysis solution and pipet several times. The solution should become viscous.
- Protein precipitation
- Add 200 µL of protein precipitation buffer to the tissue, or 1 mL to the blood. Vortex vigorously for 20 s and incubate on ice for 5 min. Small protein clumps may be visible after vortexing.
- Centrifuge at 16,000 x g for 4 min. A tight pellet containing precipitated proteins should form.
- Carefully transfer supernatant, without disturbing the pellet, to a new 1.7 mL tube.
NOTE: If pellet is not tight, consider another centrifuge at 16,000 x g for 4 min.
- DNA precipitation and rehydration
- Add 1 mL of 2-propanol to tube containing the supernatant, and gently mix by inverting. DNA should become visible as white floating substance.
- Centrifuge at 16,000 x g for 1 min. Remove the supernatant completely using a pipette.
- Add 1 mL of freshly prepared 70% ethanol, and invert tube several times. Centrifuge at 16,000 x g for 1 min at room temperature. Discard supernatant carefully.
- Repeat step 1.4.3.
- Place open tube inverted on clean absorbent paper for 10-15 min. Re-invert tube and confirm complete hydration. If needed, leave to air-dry for additional 10-15 min.
NOTE: It is crucial for pellet to dry completely. - Add 50 µL of ultra-pure water (UPW). If needed, DNA can be diluted more after quantification.
- Incubate for 1 h on heat block at 65 °C, for DNA rehydration.
- DNA quantification
NOTE: DNA was quantified using a fluorometer and designated reagents, as specified in table of materials.- Bring standards to room temperature and prepare the kit, including 0.5 mL designated tubes.
- Prepare buffer and dye mix in a 15 mL tube, according to number of samples. Add 200 µL of buffer and 1 µL of dye per sample. Include 2 samples for standards. It is recommended to calculate an additional sample in order to avoid that the buffer will not be sufficient.
- For the 2 standards, place 190 µL of the above-prepared mix in 0.5 mL tube. Add 10 µL of the standard.
- For each sample, place 199 µL of the above-prepared mix in 0.5 mL tube. Add 1 µL of the DNA.
- Read the samples. The result indicates DNA concentration of the sample.
- If samples are over the limit, dilute the DNA 1:10 with UPW, and repeat read.
2. Library preparation
NOTE: The current protocol utilizes a multiplex, PCR-based assay kit compatible with NGS sequencers. The kit contains 24 different indices each targeting conserved regions within the Vβ and the Jβ regions. This enables a one-step PCR reaction and pooling of different samples. See table of materials for details.
- Prepare DNA samples. Obtain 150 ng of DNA and add UPW for a total of 5 µL.
NOTE: It is possible to use less than 150 ng of DNA for library preparation, with minimum concentration of 10 ng/µL. - Place pipettes and tips in hood with UV light for 15 min to break down residual DNA. Meanwhile, allow different index tubes (for library preparation) and DNA polymerase to thaw on ice.
- In a biological hood, add 45 µL from the different index tubes into PCR tubes. To follow, add 0.2 µL of the DNA polymerase and the 5 µL of DNA prepared in step 2.1 into PCR tubes.
- Run PCR reaction using a pre-set program, according to the manufacturer's instructions.
3. Amplicon purification and quantification
- Use magnetic beads for removal of excess primers, nucleotides, and enzymes.
- Warm the beads at least 30 min to room temperature. Prepare enough fresh 80% ethanol for 400 µL per sample.
- Add 50 µL (IGH) or 35 µL (TCRβ) of the beads to each PCR tube and mix by pipetting 10 times. Incubate 10 min at room temperature.
- Place the mixed sample on the magnetic stand for 5 min.
- Aspirate 95 µL (IGH) or 80 µL (TCRβ) of the clear liquid and discard. Aspirate remaining liquid using a 10 µL tip.
- Add 200 µL of 80% ethanol to each sample. Incubate 30 s. Aspirate 195 µL of ethanol and discard. Aspirate remaining liquid using 10 µL tip.
- Repeat step 3.1.5. Open the caps of the tubes and let air dry for 5 min.
- Immediately after the 5 min, remove from magnetic stand and add 25 µL of elution buffer (10 mM Tris-HCl, pH 8.0). Mix by pipetting until homogeneous. Incubate at room temperature for 2 min.
- Place the tubes on the magnetic stand for 5 min. Transfer 22 µL to a new PCR tube. Measure the DNA concentration (step 1.5).
- Amplicon quantification
NOTE: The current protocol uses an automated electrophoresis machine for quality control of DNA concentration, size, and integrity. See table of materials for details.- Place reagents and screentape at room temperature for at least 30 min.
- In a new 1.7 mL micro-centrifuge tube, make a 1 ng/µL dilution of the DNA samples quantified in step 3.1.8 with UPW for a total volume of at least 3 µL.
- Vortex diluted DNA before use. Place 2 µL of buffer, together with 2 µL of the diluted DNA in the pre-labeled specialized PCR strips (provided in the kit) and place the caps. Place on shaker for 1 min, followed by spin down of PCR strips.
NOTE: Due to the viscosity of buffer, ensure excess buffer is removed from the tip before transfer to the sample tubes. - Place in the machine, the screentape and PCR strips without the caps. Open the lid of tip box inside the machine. Assign the position with appropriate labels. Start the machine.
NOTE: The output histogram should be a single peak at the desired size of the amplicons (Figure 1A). In samples of poor quality, additional peaks will be observed (Figure 1B), indicating poor bead clean-up or formation of primer dimers.
4. Next-generation sequencing
- Quantification and pooling of library
- Calculate final DNA concentration in nM, using DNA value from step 3.1.8, and the size in base pairs (bp) of the product’s peak, obtained from electrophoresis machine results (see Figure 1).
- For each sample, calculate DNA volume to take for a final concentration of 4 nM, in a final volume of 10-20 µL elution buffer.
- Prepare a new dilution mix for each sample and take 2 µL from it to a new pool mix.
NOTE: For samples that have less than 4 nM, add the maximum amount of sample possible.
- NGS run of library
NOTE: The current protocol uses a bench-top sequencer platform. See table of materials for details.- Prepare a fresh solution of 0.2 N NaOH. Add 10 µL of 0.2 N NaOH to the diluted library prepared in step 4.1.3. Add 5% PhiX to tube and vortex briefly. Spin down the tube to ensure all the solution has settled to the bottom of the tube.
- Incubate for 5 min at room temperature to denature the double stranded DNA. Add 980 μL of pre-chilled buffer from the kit to the tube containing DNA and vortex briefly.
- Place the diluted library on ice, and prepare the library as follows. For a mix of 17 pM, add 425 µL of DNA from step 4.2.2 and 575 µL of buffer. Invert library several times and spin down. Load 600 µL of the final library onto the designated cartridge, and place samples in the machine.
- Start the run following the software instructions.
5. Sequencing analysis
- Download sequencing metrics from sequencer, and verify that sequencing data is within the following ranges (specific for the kit used in the current protocol). Samples not meeting these criteria are subject to repeat run:
Cluster Density (Density (K/mm2)): 945K – 1800K
Percent of reads passing filter (Clusters PF (%)): >90%
Quality scores (%>=Q30): >85%
PhiX alignment (Aligned %): 1.5% – 10%
PhiX Error Rate (%): <1.0%
Representative Results
Herein, we describe a method for DNA isolation from intestinal tissue and blood, preparation of libraries for NGS, and basic steps of a sequencing run for immune repertoire sequencing. The run will generate fastq files, which can be further converted to fasta files for use in the international ImMunoGeneTics (IMGT)/HighV-QUEST platform. This HTS performs and manages many analyses of tens of thousands of rearranged TCRβ and IGH sequences, at the nucleotide level15. IMGT/HighV-QUEST enables analysis of different TCRs and IGH repertoires in both health and disease. This can lead to identification of new "disease-specific" clones, analysis of clonal expansion and diversity parameters, delineation of differential V(D)J usage, analysis of somatic hyper-mutations, and more. The IMGT/HighV-QUEST provides CSV files, which contain specific sequences and their abundance. Using genomic DNA as a starting material yields sequence numbers that are representative of cell numbers. Thus, if original DNA quantity is equal, percentage of T cells in a given sample can also be calculated.
We include a basic analysis from representative, autologous blood and rectal samples of a patient with IL10 receptor deficiency and history of severe infantile-onset IBD, resulting from a deleterious IL10RA mutation. Samples were assayed for both TCRβ and IGH repertoire. The intestinal TCRβ sample yielded a total 12,450 sequences, of which 9,050 sequences were unique. The blood TCRβ sample had a total 54,880 sequences, of which 35,110 were unique. In the intestinal IGH sample, a total of 49,070 sequences were obtained, of which 23,670 were unique. In the blood IGH sample, a total of 13,710 sequences were obtained, of which 13,540 were unique. All clones can be identified by their unique sequence both at the nucleotide or amino acid level. These sequences can be compared between different patients, in search for shared clones, or in the same patient between different anatomical sites (e.g., blood vs. intestine). We present for each of the samples the 5 most frequent clones (Table 1).
To quantify the degree of clonal expansion different indices can be used, including Shannon’s H, Gini-Simpson, entropy and clonality. As an example, Shannon's H, which takes into account the number of unique sequences (richness of the repertoire) and how evenly they are distributed was found to be decreased in the patient's intestinal IGH vs. blood (8.3 vs. 9.5), suggesting clonal expansion of B cells in the inflamed gut.
For a broad overview of the repertoire, Treemap images (www.treemap.com) were generated (Figure 2). Each colored square represents a different clone, and the size correlates with its frequency. These Treemap images demonstrate clonal expansion in the intestinal IGH repertoire compared with the blood. In contrast, in the TCRβ repertoire, marked clonal expansion is observed in the blood, in comparison to autologous intestine.
At the gene level, NGS provides information regarding V- D- and J- usage at the level of either gene, family, or allele, as shown in Table 1. Moreover, specific V(D)J combinations can be inferred from the data for TCRβ and IGH repertoires (Figure 3A,B), which can reveal differential gene usage patterns in various conditions. Biophysical properties of the CDR3 region such as length (Table 1) or hydrophobicity can also be analyzed. Importantly, CDR3 length distribution is altered in different immune-mediated disorders10,16,17. For example, in IL10R deficient patients, blood-derived T cells, but not B cells, have shorter CDR3 length, and differential hydrophobicity, in comparison to healthy controls7.
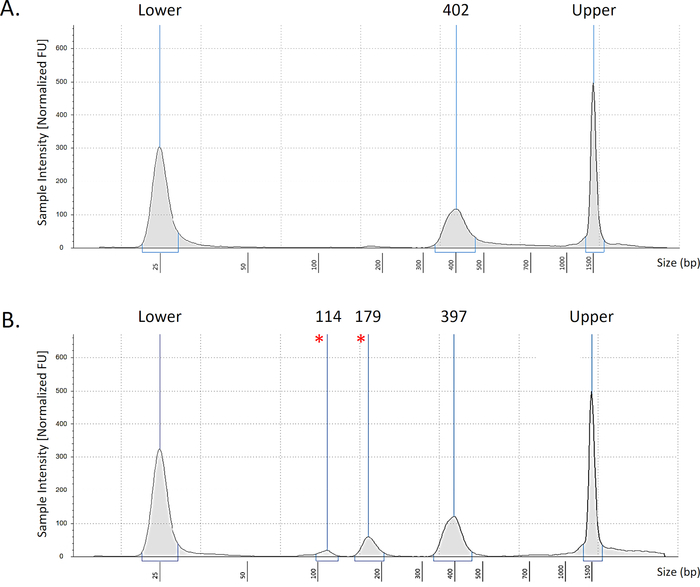
Figure 1: Representative bioanalyzer data. Representative images of intestinal TCRβ samples showing optimal results (A) with the desired peak at 400 bp. An example of a low-quality library (B) with additional peaks at 179bp and 114bp (marked *). Peaks seen at ends are upper and lower markers of the electrophoresis strip. Please click here to view a larger version of this figure.
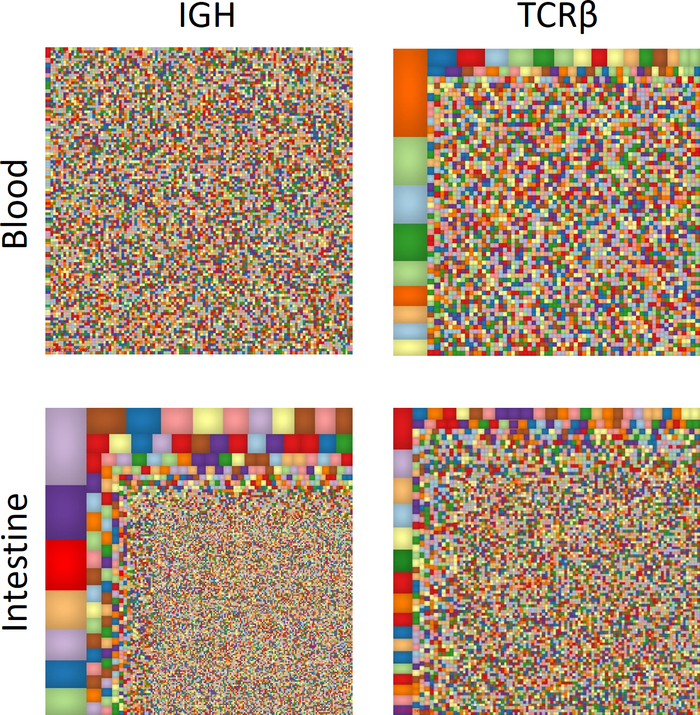
Figure 2: Overview of T and B cell immune repertoire. Treemap diagrams of TCRβ and IGH blood and intestinal samples. Each colored square represents a different clone, and the size correlates with its frequency. Please click here to view a larger version of this figure.
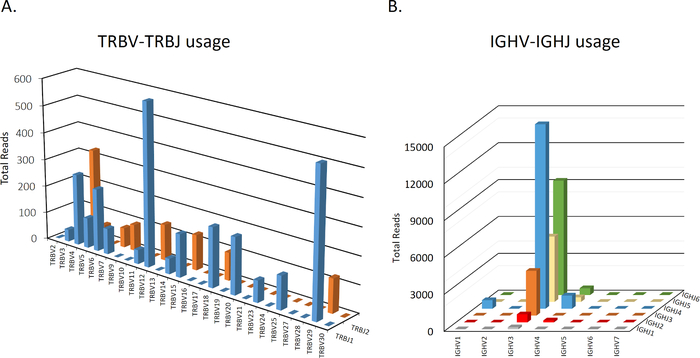
Figure 3: Representative graphs of TCRβ and IGH V-J usage. A representative intestinal sample depicting total number of sequences for each V-J combination in TCRβ (A) or IGH (B). Data is not shown for D usage. Please click here to view a larger version of this figure.
Discussion
Changes in abundance and function of B and T lymphocytes are often encountered in different malignancies18, chronic inflammatory disorders (e.g., ulcerative colitis and rheumatoid arthritis)10,19, and in various immunodeficiencies17,20. The current method utilizes NGS to facilitate an in-depth view of TCR and BCR repertoires, enabling detection of subtle changes in T and B cell clonality, sharing of clones, V(D)J gene usage, and information on the degree of somatic hyper-mutations in the case of B cells.
The method of DNA isolation was described for intestinal biopsies and blood samples. However, with modifications of lysis and DNA extraction, the method can be applied to other tissues such as tumors, lymph nodes, synovial fluid, etc. It is important during library preparation not to cross-contaminate the different primer sets. At bead clean-up, care should be taken to leave tubes on the magnet at all times of incubation. For DNA samples of low concentrations, stocks can be concentrated at 65 °C for 10-20 minutes. Moreover, amplicons can be eluted from beads in as little as 20 µL elution buffer.
Different techniques used in the past to characterize the landscape of T and B cell composition provided only a superficial overview of the immune repertoire. One of the biggest advantages of NGS for immune repertoire analysis is the ability to identify unique clones, and consequently track them in different anatomical sites (e.g., intestine vs. blood vs. lymph node) or in different individuals. The clinical implications of such a technique are significant, and go beyond enhancing the understanding of the role of T or B cells in different immune-mediated diseases or malignancies21. In oncology, NGS is used to identify specific clones that can be predictive biomarkers for the patients who will most likely benefit from current immunotherapies22,23,24. In leukemia, NGS is used to confirm full eradication of cancerous cells by detection of residual leukemic clones that remain in a patient after treatment25.
One of the main limitations of immune repertoire analysis of whole tissue or blood samples is the inability to identify repertoire changes in less frequent populations. For example, if regulatory T cells, which comprise only a few percentages of total CD4+ T cells, express a unique repertoire profile, this would be missed if conducting these studies on whole blood. This problem could be addressed by conducting these studies on sorted immune populations. Alternatively, in recent years technology has developed to facilitate coupling of single cell RNA data with TCR or IGH repertoire profiles26. This provides another level of functionality of the T or B cells, since the transcriptional landscape of each clone can be characterized. As an example, Zemmour et al. used scRNAseq TCRseq to demonstrate that regulatory T cells from human blood, display broad heterogeneity with an activated subpopulation that is transcriptionally related to conventional T cells27. Similarly, in patients with hepatocellular carcinoma this method identified 11 distinct T cell subsets that infiltrate the tumor, each one with a unique transcriptional and repertoire profile28. Studies like these will be helpful especially when studying rare populations, and will provide important functional information of specific clones.
NGS of immune repertoire is a new research field that provides novel insights about adaptive immune function and the role of T and B cells in various diseases. In addition, it is rapidly entering the clinical world in different disciplines, by tracking disease associated clonotypes. There are currently several available kits for immune repertoire. These are based on similar methodology, regardless of whether they use RNA or DNA as the source. They use similar techniques for calibration, bias corrections, and analysis tools; and thus need to be properly chosen according to specific demands. One of the great advantages of the current kit is that it is a ready-to-use and optimized for human immune repertoire, without need for calibration. In the future we will see more studies using repertoire features as biomarkers for different disease, potentially also leading to development of targeted therapies against these clones. We believe researchers should consider applying these methods for studying the adaptive immune landscape in different disorders.
Disclosures
The authors have nothing to disclose.
Acknowledgements
None.
Materials
| 2-propanol | Sigma | I9516-500ML | |
| 1.7 mL micro-centrifuge tubes | Axygen | 8187631104051 | |
| 15 mL centrifuge tubes | Greiner | 188261 | |
| Absolute ethanol | Merck | 1.08543.0250 | |
| Amplitaq Gold | Thermo Fisher | N8080241 | |
| AMPure XP Beads | Beckman Coulter | A63881 | |
| Heat block | Bioer | Not applicable | |
| High Sensitivity D1000 Sample Buffer | Agilent | 5067-5603 | For Tapestation |
| High Sensitivity D1000 ScreenTape | Agilent | 5067-5584 | For Tapestation. Tubes sold seperately |
| Lymphotrack Assay kit | Invivoscribe | TRB: 70-91210039 IGH: 70-92250019 | Each includes 24 indexes |
| MiSeq Reagent Kit v2 (500 cycle) | Illumina | MS-102-2003 | Includes standard flow cell type and all reagents required |
| MiSeq Sequencer | Illumina | SY-410-1003 | |
| PCR strips | 4titude | 4ti-0792 | |
| Proteinase K | Invitrogen | EO0491 | |
| Qubit 4 Fluorometer | Thermo Fisher | Q33226 | |
| Qubit dsDNA HS Assay Kit | Thermo Fisher | Q32854 | Includes buffer, dye, standards, and specialized tubes |
| Shaker | Biosan | Not applicable | |
| Tapestation 2100 Bioanalyzer | Agilent | G2940CA | |
| ultra pure water | Bio-lab | 7501 | |
| Wizard DNA isolation kit | Promega | A1120 | Includes cell lysis solution, nuclei lysis solution, and protein precipitation buffer |
References
- Bassing, C. H., Swat, W., Alt, F. W. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 109, 45-55 (2002).
- Roth, D. B. V(D)J Recombination: Mechanism, Errors, and Fidelity. Microbiology Spectrum. 2 (6), (2014).
- Heather, J. M., Ismail, M., Oakes, T., Chain, B. High-throughput sequencing of the T-cell receptor repertoire: pitfalls and opportunities. Brief Bioinformatics. 19 (4), 554-565 (2018).
- Pabst, O., Hazanov, H., Mehr, R. Old questions, new tools: does next-generation sequencing hold the key to unraveling intestinal B-cell responses. Mucosal Immunology. 8 (1), 29-37 (2015).
- Bashford-Rogers, R. J. M., Smith, K. G. C., Thomas, D. C. Antibody repertoire analysis in polygenic autoimmune diseases. Immunology. 155 (1), 3-17 (2018).
- Lee, Y. N., et al. Characterization of T and B cell repertoire diversity in patients with RAG deficiency. Science Immunology. 1 (6), (2016).
- Werner, L., et al. Alterations in T and B Cell Receptor Repertoires Patterns in Patients With IL10 Signaling Defects and History of Infantile-Onset IBD. Frontiers Immunology. 11, 109 (2020).
- Zhang, J., et al. Immune receptor repertoires in pediatric and adult acute myeloid leukemia. Genome Medicine. 11 (1), 73 (2019).
- Chapman, C. G., et al. Characterization of T-cell Receptor Repertoire in Inflamed Tissues of Patients with Crohn’s Disease Through Deep Sequencing. Inflammatory Bowel Diseases. 22 (6), 1275-1285 (2016).
- Werner, L., et al. Altered T cell receptor beta repertoire patterns in pediatric ulcerative colitis. Clinical and Experimental Immunology. 196 (1), 1-11 (2019).
- Bashford-Rogers, R. J. M., et al. Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature. 574 (7776), 122-126 (2019).
- Wu, J., et al. Expanded TCRbeta CDR3 clonotypes distinguish Crohn’s disease and ulcerative colitis patients. Mucosal Immunology. 11 (5), 1487-1495 (2018).
- Rosati, E., et al. Identification of disease-associated traits and clonotypes in the T-cell receptor repertoire of monozygotic twins affected by inflammatory bowel diseases. Journam of Crohn’s and Colitis. , (2019).
- Allez, M., et al. T cell clonal expansions in ileal Crohn’s disease are associated with smoking behaviour and postoperative recurrence. Gut. 68 (11), 1961-1970 (2019).
- Li, S., et al. IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nature Communications. 4, 2333 (2013).
- H, I. J., et al. Strategies for B-cell receptor repertoire analysis in primary immunodeficiencies: from severe combined immunodeficiency to common variable immunodeficiency. Frontiers Immunology. 6, 157 (2015).
- Ghraichy, M., Galson, J. D., Kelly, D. F., Truck, J. B-cell receptor repertoire sequencing in patients with primary immunodeficiency: a review. Immunology. 153 (2), 145-160 (2018).
- Zhuang, Y., et al. Application of immune repertoire sequencing in cancer immunotherapy. International Immunopharmacology. 74, 105688 (2019).
- Liu, X., et al. T cell receptor beta repertoires as novel diagnostic markers for systemic lupus erythematosus and rheumatoid arthritis. Annual Rheumatic Diseases. 78 (8), 1070-1078 (2019).
- Wong, G. K., Heather, J. M., Barmettler, S., Cobbold, M. Immune dysregulation in immunodeficiency disorders: The role of T-cell receptor sequencing. Journal of Autoimmunity. 80, 1-9 (2017).
- Delhalle, S., Bode, S. F. N., Balling, R., Ollert, M., He, F. Q. A roadmap towards personalized immunology. NPJ System Biology and Applications. 4, 9 (2018).
- Laubli, H., et al. The T cell repertoire in tumors overlaps with pulmonary inflammatory lesions in patients treated with checkpoint inhibitors. Oncoimmunology. 7 (2), 1386962 (2018).
- Hogan, S. A., et al. Peripheral Blood TCR Repertoire Profiling May Facilitate Patient Stratification for Immunotherapy against Melanoma. Cancer Immunology Research. 7 (1), 77-85 (2019).
- Aversa, I., Malanga, D., Fiume, G., Palmieri, C. Molecular T-Cell Repertoire Analysis as Source of Prognostic and Predictive Biomarkers for Checkpoint Blockade Immunotherapy. International Journal of Molecular Sciences. 21 (7), (2020).
- Hirsch, P., et al. Precision and prognostic value of clone-specific minimal residual disease in acute myeloid leukemia. Haematologica. 102 (7), 1227-1237 (2017).
- De Simone, M., Rossetti, G., Pagani, M. Single Cell T Cell Receptor Sequencing: Techniques and Future Challenges. Frontiers Immunology. 9, 1638 (2018).
- Zemmour, D., et al. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nature Immunology. 19 (3), 291-301 (2018).
- Zheng, C., et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 169 (7), 1342-1356 (2017).

