Organic Solvent-Based Protein Precipitation for Robust Proteome Purification Ahead of Mass Spectrometry
Summary
The present protocol describes solvent-based protein precipitation under controlled conditions for robust and rapid recovery and purification of proteome samples prior to mass spectrometry.
Abstract
While multiple advances in mass spectrometry (MS) instruments have improved qualitative and quantitative proteome analysis, more reliable front-end approaches to isolate, enrich, and process proteins ahead of MS are critical for successful proteome characterization. Low, inconsistent protein recovery and residual impurities such as surfactants are detrimental to MS analysis. Protein precipitation is often considered unreliable, time-consuming, and technically challenging to perform compared to other sample preparation strategies. These concerns are overcome by employing optimal protein precipitation protocols. For acetone precipitation, the combination of specific salts, temperature control, solvent composition, and precipitation time is critical, while the efficiency of chloroform/methanol/water precipitation depends on proper pipetting and vial manipulation. Alternatively, these precipitation protocols are streamlined and semi-automated within a disposable spin cartridge. The expected outcomes of solvent-based protein precipitation in the conventional format and using a disposable, two-stage filtration and extraction cartridge are illustrated in this work. This includes the detailed characterization of proteomic mixtures by bottom-up LC-MS/MS analysis. The superior performance of SDS-based workflows is also demonstrated relative to non-contaminated protein.
Introduction
Proteome analysis by mass spectrometry has become increasingly rigorous, owing to the enhanced sensitivity, resolution, scan speed, and versatility of modern MS instruments. MS advances contribute to greater protein identification efficiency and more precise quantitation1,2,3,4,5. With improved MS instrumentation, researchers demand a correspondingly consistent front-end sample preparation strategy capable of quantitative recovery of high-purity proteins in minimal time across all stages of the workflow6,7,8,9,10,11. To accurately reflect the proteome status of a biological system, proteins must be isolated from the native sample matrix in an efficient and unbiased fashion. To this end, including a denaturing surfactant, such as sodium dodecyl sulfate (SDS), ensures efficient protein extraction and solubilization12. However, SDS strongly interferes with electrospray ionization, causing severe MS signal suppression if not properly eliminated13.
Various SDS depletion strategies are available for subsequent proteome analysis, such as the retention of proteins above a molecular weight cutoff filter contained within disposable spin cartridges14,15,16. The filter-aided sample preparation method (FASP) is favored as it effectively depletes SDS below 10 ppm, facilitating optimal MS. However, protein recovery with FASP is variable, which prompted the exploration of other techniques. Chromatographic approaches that selectively capture protein (or surfactant) have evolved into various convenient cartridges or bead-based formats17,18,19,20,21. Given these simple and (ideally) consistent strategies to protein purification, the classical approach of protein precipitation with organic solvents is often overlooked as a promising approach to protein isolation. While solvent precipitation is shown to deplete SDS below critical levels successfully, protein recovery has been a longstanding concern of this approach. Multiple groups have observed a protein recovery bias, with unacceptably low precipitation yields as a function of protein concentration, molecular weight, and hydrophobicity22,23. Due to the diversity of precipitation protocols reported in the literature, standardized precipitation conditions were developed. In 2013, Crowell et al. first reported the dependence of ionic strength on the precipitation efficiency of proteins in 80% acetone24. For all proteins examined, the addition of up to 30 mM sodium chloride was shown to be essential to maximize yields (up to 100% recovery). More recently, Nickerson et al. showed that the combination of even higher ionic strength (up to 100 mM) with elevated temperature (20 °C) during acetone precipitation gave near quantitative recovery in 2-5 min25. A slight drop in the recovery of low molecular weight (LMW) proteins was observed. Therefore, a subsequent report by Baghalabadi et al. demonstrated the successful recovery of LMW proteins and peptides (≤5 kDa) by combining specific salts, particularly zinc sulfate, with a higher level of organic solvent (97% acetone)26.
While refining the precipitation protocol lends a more reliable protein purification strategy for MS-based proteomics, the success of conventional precipitation relies heavily on user technique. A primary goal of this work is to present a robust precipitation strategy that facilitates the isolation of the protein pellet from the contaminating supernatant. A disposable filtration cartridge was developed to eliminate pipetting by isolating aggregated protein above a porous PTFE membrane filter27. MS-interfering components in the supernatant are effectively removed in a short, low-speed centrifugation step. The disposable filter cartridge also offers an interchangeable SPE cartridge, which facilitates subsequent sample clean-up following resolubilization and optional protein digestion, ahead of mass spectrometry.
A series of recommended proteome precipitation workflows are presented here, including modified acetone and chloroform/methanol/water28 protocols, in a conventional (vial-based) and a semi-automated format in a disposable two-state filtration and extraction cartridge. The resulting protein recoveries and SDS depletion efficiencies are highlighted, together with bottom-up LC-MS/MS proteome coverage, to demonstrate the expected outcome from each protocol. The practical benefits and drawbacks associated with each approach are discussed.
Protocol
1. Material considerations and sample pre-preparation
- Use only high purity solvents (acetone, chloroform, methanol) (>99.5%) and chemicals, free of excess moisture.
- Prepare sodium chloride and zinc sulfate solutions (1 M) in water.
NOTE: Salt solutions can be stored indefinitely at room temperature, as long as they are free of contaminant or microbial growth. - Use the smallest polypropylene (PP) microcentrifuge vial sufficient to retain the required volume of sample and solvents to induce precipitation.
- Ensure that the SDS concentration in the sample to be precipitated is no greater than 2% (w/v). If SDS is higher, dilute the sample with water.
- Ensure a protein concentration between 0.01 and 10 g/L for optimal precipitation efficiency.
NOTE: The optimal mass for precipitation ranges between 1-100 µg of protein. - Ensure all solvents and solutions are free of particulate matter before use. Perform either a filtration (<0.5 µm) or centrifugation step (10,000 x g for 1 min at room temperature) to remove undissolved particulates.
- If disulfide bond reduction and alkylation are required, conduct these steps prior to protein precipitation. Excess reducing and alkylating reagents will be removed through the precipitation process.
- Precipitate the proteins by selecting and performing one of the protocols (steps 2, 3, 4, or 5).
2. Rapid (vial-based) protein precipitation with acetone
- Pipet 90 µL of (particulate-free) protein or proteome solution into a PP microcentrifuge tube. Then, add 10 µL of 1 M aqueous NaCl.
NOTE: If the ionic strength of the proteome extract already exceeds 100 mM, no additional salt is necessary. - Pipet 400 µL of acetone into the sample. Cap the vial and tap the vial gently to combine the solvents. Vigorous mixing is not required.
NOTE: The volume of protein, salt, and acetone can be increased so long as the relative ratio of each is maintained. - Allow the vial to incubate at room temperature, undisturbed, for a minimum of 2 min.
NOTE: Longer incubations, including those at reduced temperature (e.g., conventional acetone precipitation employs overnight precipitation in the freezer), may result in the formation of larger (visible) aggregated protein particulates (Figure 1A), which generally do not improve total protein recovery. - Following incubation, place samples in a centrifuge, noting the orientation of the vial. Spin for a minimum of 2 min, at 10,000 x g or higher at room temperature.
- Uncap the vial and gently decant the supernatant by slowly inverting the vial to a waste container. Touch the inverted vial to a paper towel to draw residual solvent from the vial.
CAUTION: Waste solvents should be retained and discarded as per appropriate protocols. - For SDS-containing samples, dispense 400 µL of fresh acetone, being careful not to disturb the pellet.
NOTE: Step 2.6 is optional.- Immediately centrifuge the sample (10,000 x g or higher for 1 min at room temperature), placing the vial into the rotor in the same orientation as the initial spin. Decant the wash solvent as described in step 2.5.
- Allow the sample to fully dry with the cap open (~1 min). Recap the vial and proceed with pellet solubilization (step 6).
3. Precipitation of low molecular weight (LMW) peptides (ZnSO4 + acetone)
- Dispense 54 µL of proteome extract to a 2 mL PP vial, and then add 6 µL of 1 M ZnSO4.
NOTE: Optimal recovery of LMW peptides (≤5 kDa) is obtained by adding acetone to a final 97% by volume. Assuming a 2 mL PP vial, the maximal initial sample volume is 54 µL. - Add 1940 µL of acetone to a final 97% by volume. Swirl gently to mix, and let stand undisturbed on the benchtop for a minimum of 2 min.
- Centrifuge (10,000 x g for 1 min at room temperature) and remove the supernatant by inverting the vial, and then touching the vial to a paper towel.
- For SDS-containing samples, dispense 400 µL of fresh acetone, being careful not to disturb the pellet.
NOTE: Step 3.4 is optional.- Immediately centrifuge the sample as per step 3.3, placing the vial into the rotor in the same orientation as the initial spin. Decant the wash solvent as described in step 3.3.
- Re-solubilize the resulting dry pellet in an aqueous solvent with brief vortexing or sonication (~5 min).
4. Protein precipitation by chloroform/methanol/water (CMW)
- Dispense 100 µL of the protein or proteome solution into a PP vial. Add 400 µL of methanol, followed by 100 µL of chloroform. Cap the vial and vortex briefly to mix.
NOTE: For CMW precipitation, 1.5 mL vials with narrow bottoms are preferred (Figure 2A).
CAUTION: Chloroform solvent should be handled in an appropriate ventilation hood. All solvents that contact chloroform should be treated as halogenated waste when disposed of. - Quickly dispense 300 µL of water directly into the center of the vial. Cap the vial. Allow the sample to sit on the benchtop undisturbed for 1 min.
NOTE: The solution will immediately appear cloudy white. Avoid mixing the vial following the addition of water. - Place the PP vial in a centrifuge and spin for a minimum of 5 min (10,000 x g or higher at room temperature).
NOTE: Once centrifuged, two visible solvent layers will form (top layer = methanol/water; bottom = chloroform). A solid protein pellet forms at the solvent interface (Figure 2A). - Using a large (1 mL) micropipette tip, and holding the vial at ~45°, remove ~700 µL of the solvent from the upper layer at a uniform rate.
- Use a smaller (200 µL) micropipette tip to continue removing the upper solvent layer from the ~45° tilted vial. Pipet in one continuous motion until the upper solvent layer forms a bead in the vial.
- Add 400 µL of fresh methanol to the sample vial, without disturbing the pellet, by dispensing the solvent down the side of the vial.
- Cap the vial. Combine the solvent layers by gently rocking the vial to swirl the solvents together.
NOTE: It is essential to avoid disrupting the pellet. Do not vortex the vial. - Noting the orientation of the vial in the rotor, centrifuge for a minimum of 10 min (10,000 x g at room temperature). The protein pellet adheres to the bottom of the vial (Figure 2B).
- Tip the vial at 45°, with the pellet facing down. Place the pipette tip along the upper edge of the vial and remove the supernatant with a 1 mL micropipette tip at a slow but continuous rate. Retain ~20 µL of solvent in the vial.
- Wash the protein pellet for SDS-containing samples by slowly dispensing 400 µL of fresh methanol. Do not vortex the vial.
- Proceed directly with centrifugation (10,000 x g for 2 min at temperature), placing the vial into the rotor with the same orientation as the initial spin.
- Remove the solvent, as per step 4.9. Allow the sample to air dry in a fumehood until the residual solvent evaporates.
- Consult the recommended resolubilization procedures in step 6.
5. Protein precipitation using a disposable filtration cartridge
NOTE: Each solvent-based precipitation protocol described in steps 2-5 can be performed in a two-stage filtration and extraction cartridge (see Table of Materials).
- With the plug attached to the upper filtration cartridge (Figure 3A), dispense the desired volume of the extracted proteome, salt, and solvent as outlined in one of the three options below.
- (Option 1) For protein precipitation with acetone, combine 90 µL of protein or proteome solution, 10 µL of 1 M aqueous NaCl, and 400 µL of acetone. Incubate for a minimum of 2 min on the benchtop.
NOTE: A visible precipitate will develop for concentrated protein samples (1 g/L) (Figure 3B). - (Option 2) For LMW peptide precipitation, combine 15 µL of the sample, 1.5 µL of 1 M ZnSO4, and 485 µL of acetone. Incubate for a minimum of 2 min on the benchtop.
NOTE: A salt concentration of 90 mM in the aqueous sample will not impact recovery relative to the 100 mM recommended in step 3. - (Option 3) For CMW precipitation, add 50 µL of proteome extract, 200 µL of methanol, and 50 µL of chloroform. Cap the vial and briefly vortex to combine.
- Quickly dispense 150 µL of water directly into the center of the vial. Incubate for 1 min on the benchtop.
- (Option 1) For protein precipitation with acetone, combine 90 µL of protein or proteome solution, 10 µL of 1 M aqueous NaCl, and 400 µL of acetone. Incubate for a minimum of 2 min on the benchtop.
- Centrifuge for 2 min at 2,500 x g at room temperature with the plug still attached to the filtration cartridge.
- Invert the cartridge, and then unscrew and remove the plug from the cartridge base.
- Place the filtration cartridge in a clean vial and return to the centrifuge. Spin for 3 min at 500 x g at room temperature. Discard the flow-through solvent from the lower vial.
NOTE: If any solvent remains in the upper filtration cartridge, return to the centrifuge and perform an additional spin. - Wash the protein pellet by adding 400 µL of acetone to the filtration cartridge (for CMW precipitation, step 5.1.3, add 400 µL of methanol).
- Centrifuge for 3 min at 500 x g at room temperature or until no solvent remains in the upper cartridge.
- Re-solubilize the precipitation pellet as described in step 6.
6. Resolubilization of protein pellet
- Wet the membrane at the base of the filtration cartridge by dispensing 2-5 µL of isopropanol directly to the membrane immediately before the resolubilization protocols described below.
- Follow one of the following resolubilization methods.
- (Option 1) Add a minimum of 20 µL of aqueous buffer containing ≥2% SDS to the filtration cartridge. Cap and vortex vigorously (~1 min). Alternatively, sonicate (>10 min) to disperse the protein pellet.
- Heat the sample at 95 °C for 5 min). Repeat the mixing step after heating.
NOTE: Laemmli gel loading buffer can re-solubilize the protein pellet. However, SDS-containing samples are incompatible with trypsin digestion and reversed-phase LC and MS.
- Heat the sample at 95 °C for 5 min). Repeat the mixing step after heating.
- (Option 2) Prepare a solution of 80% (v/v) formic acid in water. Prechill the acid solution (-20 °C), as well as the filtration cartridge containing precipitated protein.
- Dispense 50 µL of cold formic acid into the cartridge; cap and vortex for 30 s. Return to the freezer (-20 °C) for 10 min.
- Vortex the cartridge again for 30 s. Then, repeat the chilling and mixing cycle one more time (10 min, -20 °C, 30 s vortex).
- Add water to a final 500 µL, diluting the formic acid to 8%.
NOTE: The cold formic acid protocol is incompatible with subsequent trypsin digestion but is compatible with LC-MS.
- (Option 3) Add 50 µL of freshly prepared 8 M urea in water to the filtration cartridge. Sonicate for 30 min.
- Allow the cartridge to incubate on the benchtop for 1 h (up to overnight).
- Dilute the 8 M urea a minimum 5-fold with water or appropriate buffer.
NOTE: Once diluted, the urea solubilization protocol is compatible with subsequent trypsin digestion, as well as LC-MS.
- (Option 1) Add a minimum of 20 µL of aqueous buffer containing ≥2% SDS to the filtration cartridge. Cap and vortex vigorously (~1 min). Alternatively, sonicate (>10 min) to disperse the protein pellet.
7. Protein digestion
- For bottom-up MS analysis, subject the re-solubilized proteins to enzymatic digestion using one of the two methods mentioned below.
- (Option 1) For formic acid resolubilization, reduce the initial volume of 80% formic acid in step 6.2.2.1 to 25 µL. In step 6.2.2.3, use 375 µL of water to dilute the formic acid to 5% (v/v).
- Dispense pepsin into the cartridge at an approximate protein to enzyme ratio of 50:1. With a plug attached to the filtration cartridge, incubate the sample overnight at room temperature.
- (Option 2) For resolubilization in urea, ensure a pH between 8 and 8.3 with the inclusion of 100 mM of Tris or ammonium bicarbonate in step 6.2.3.2.
- Add trypsin at an approximate protein to enzyme mass ratio of 50:1. With a plug attached to the cartridge, incubate the sample overnight in a warm water bath at 37 °C.
- Terminate the digestion by acidifying the solution with 10% TFA to a final 1%.
- Recover the pepsin- or trypsin-digested protein by removing the plug from the base of the filter and centrifuging the cartridge contained within a clean vial (2 min, 5000 x g, room temperature).
8. SPE clean-up
NOTE: For additional sample desalting following digestion or solvent exchange, the sample can be subject to reversed-phase clean-up as described.
- Prime an SPE cartridge (see Table of Materials) by passing 300 µL of methanol (2 min, 400 x g) followed by 300 µL of 5% acetonitrile/0.1% of TFA (2 min, 400 x g).
- Connect the primed SPE cartridge to the base of the filtration cartridge containing re-solubilized or digested protein.
- Spin the protein through the SPE cartridge (5 min, 800 x g at room temperature). If solvent remains in the upper cartridge, return the cartridge to the centrifuge and repeat the spin.
NOTE: Passing the sample through the SPE cartridge a second time may improve recovery. - Add 300 µL of 5% acetonitrile/0.1% of TFA in water to the cartridge. To wash, flow through the SPE cartridge (2 min, 2000 x g). Discard the flow-through.
- For LMW proteins or digested peptides, elute the sample by flowing 300 µL of 50% acetonitrile/0.1% TFA (5 min, 2500 x g).
- For intact proteins, follow step 8.5 with an additional elution step using 300 µL of 75% acetonitrile/0.1% TFA. Combine the two resulting extracts.
NOTE: Step 8.6 is optional.
Representative Results
Figure 4 summarizes the expected SDS depletion following vial-based or cartridge-facilitated precipitation of proteins in a disposable filter cartridge using acetone. Conventional overnight incubation (-20 °C) in acetone is compared to the rapid acetone precipitation protocol at room temperature (step 2), as well as CMW precipitation (step 4). Residual SDS was quantified by the methylene blue active substances (MBAS) assay29. Briefly, 100 µL sample was combined with 100 µL MBAS reagent (250 mg methylene blue, 50 g sodium sulfate, 10 mL sulfuric acid, diluted in water to 1.0 L), followed by the addition of 400 µL chloroform and absorbance measurement of the organic layer at 651 nm on a UV/Vis spectrophotometer. All approaches reduce SDS to permit optimal MS analysis.
Quantitative and reproducible protein recovery is achieved following rapid acetone precipitation and CMW precipitation, as seen in Figure 5 through SDS PAGE analysis of a processed yeast total cell lysate. Precipitation in a disposable filtration cartridge eliminates the need to carefully pipet the SDS-containing supernatant while retaining the aggregated proteins above a membrane filter. Consistent recovery is obtained with all precipitation protocols, with no visible bands detected in the supernatant fractions across three independent replicates.
Figure 6 quantifies the expected yields, including the resolubilization of precipitated protein pellets using cold formic acid (step 6). CMW precipitation affords quantitative recovery by carefully preserving the pellet in a vial-based approach (step 5), which equals that obtained using the cartridge (100 ± 4% vs. 101 ± 3%, respectively). Recovery of acetone-precipitated protein pellets benefits from a filtration cartridge, with a 15-20 % improvement in yield observed. In vials, isolation of the acetone supernatant from the aggregated protein essentially relies on the adherence of the pellet to the PP tube surface; the filtration cartridge eliminates this concern as the filter ensures high recovery of precipitated protein without pipetting.
To efficiently recover LMW proteins and peptides, the acetone precipitation protocol is modified by substituting NaCl for ZnSO4 and raising the solvent percentage to 97%. Combining this specific salt and higher levels of organic solvent are required for the high recovery of LMW proteins and peptides26. As seen in Figure 7, cartridge-based protein precipitation demonstrates superior recovery of a pepsin-digested sample of bovine plasma relative to vial-based precipitation. The disposable spin cartridge can recover over 90% of LMW peptides. More significant differences in yield are noted in the cartridge when employing NaCl, confirming the importance of salt type to maximize yield. Including ZnSO4 as opposed to NaCl results in an aggregated protein pellet that is more readily trapped by the spin cartridge filter.
To assess the efficacy of precipitating proteins over a wide dynamic range, a mixture of three standard proteins was processed: β-galactosidase (β-gal) from E. coli, cytochrome c (Cyt c) from bovine, and enolase (Eno) from S. cerevisiae. The mass ratio of β-gal:Cyt c:Eno was 10,000:10:1. Samples initially contained 2% of SDS prior to cartridge-based precipitation (step 5) and were re-solubilized and digested with trypsin (steps 6 and 7). Samples prepared in vials acted as a control, having no SDS and omitting the precipitation. All samples were subject to equivalent SPE clean-up (step 8). Bottom-up MS was conducted, with MS/MS spectra searched against a combined database containing all proteins from the three species involved (see Table of Materials for instrument and software platforms). A peptide false discovery rate of 1% was employed. All three proteins were identified by MS, with 666, 28, and 35 unique peptides for β-gal, Cyt c, and Eno, respectively. Figure 8 quantifies the relative ratio (peptide peak intensity) from each sample, with a ratio above 1 reflecting a higher peptide abundance for samples processed in the disposable filter cartridges. The results demonstrate the benefits of incorporating SDS into a proteomics workflow, minimizing protein loss (e.g., from potential adsorption to the sample vial), and maximizing peptide yields.
Bovine liver was procured at a local grocery store. The proteins were isolated by extracting the tissue with a solution of 1% SDS. Subsequently, the recovered proteome was precipitated, re-solubilized (urea), and digested with trypsin, all within a disposable cartridge. Bottom-up LC-MS/MS was conducted, resulting in the identification of an average of ~8,000 proteins (~30,000 peptides). False discovery rates of 0.5% and 1.0% for peptide spectra and protein groups, was employed, searching the bovine database. The technical reproducibility of this cartridge-based workflow is assessed through overlapping protein identifications. The replicate MS injections of a common digested sample achieves on average 78 ± 0.5% overlap with the identified proteins. By comparison, samples independently prepared in discrete cartridges achieved 76 ± 0.5% overlap. These data suggest that the contribution of sample preparation toward the total variability of the analysis is minor, relative to that already contributed by the LC-MS instrumental approach. The bovine proteins identified from three technical replicates (processed independently in three disposable cartridges) were further characterized concerning their molecular weight, hydrophobicity, and isoelectric point, shown in Figure 9. A two-way ANOVA could not determine statistical differences in the identified proteomes across the technical replicates. Finally, Figure 10 compares the number of identified peptides per protein across the three replicate sample preparations. The correlation coefficients in these graphs (0.94-0.95) demonstrate the high consistency of the sample preparation approach for bottom-up MS analysis.
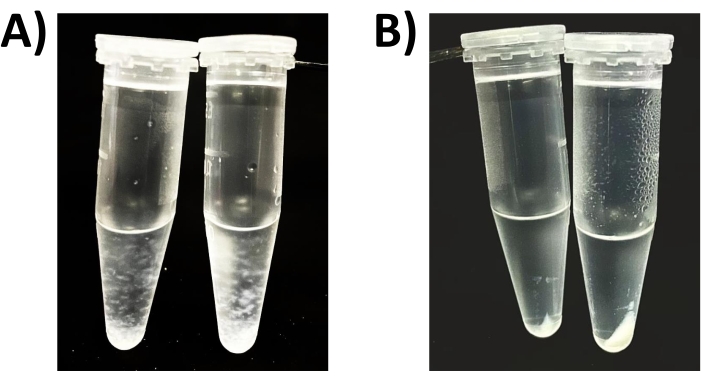
Figure 1: Acetone-precipitated proteins. Samples containing 100 and 1,000 µg of protein combined with 100 mM NaCl and precipitated with 80% acetone (A) following 5 min precipitation time and (B) following precipitation and subsequent centrifugation. Please click here to view a larger version of this figure.
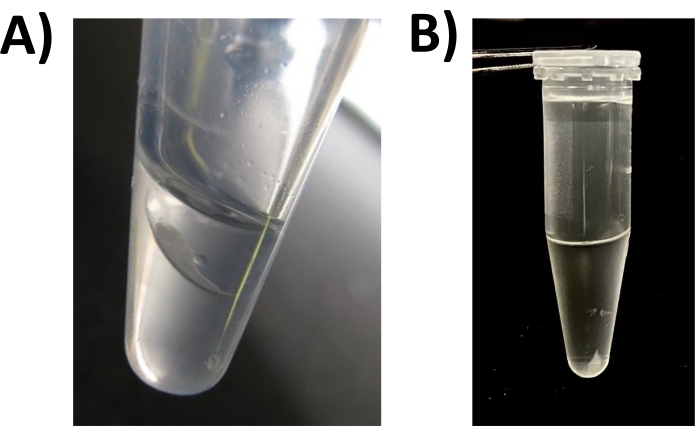
Figure 2: Protein precipitation by chloroform/methanol/water. A sample containing 50 µg of protein precipitated as per step 4. (A) Immediately following step 4.4. (B) Immediately following step 4.8. Please click here to view a larger version of this figure.
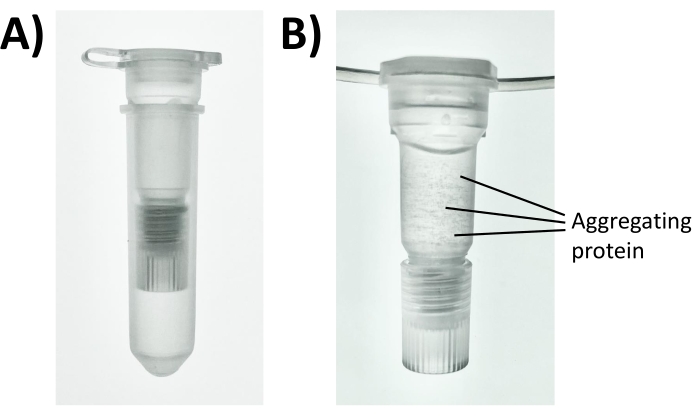
Figure 3: Photos of a disposable two-stage filtration and extraction cartridge for protein precipitation. A sample containing 100 µg protein was combined with 100 mM of NaCl and 80% acetone in (A) the assembled filtration and SPE cartridge and (B) precipitated for 5 min until protein aggregates became visible. Please click here to view a larger version of this figure.
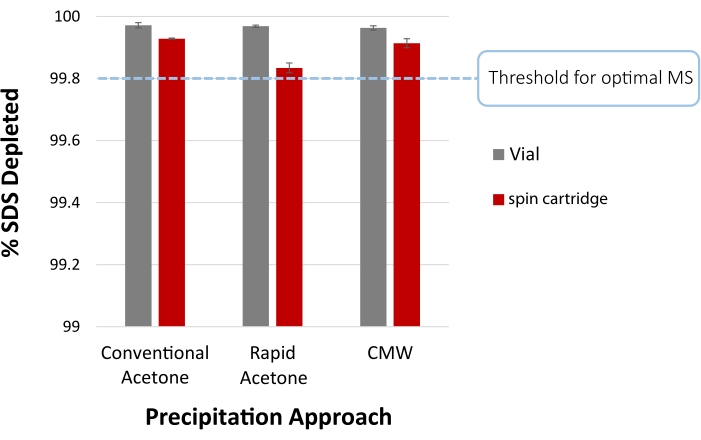
Figure 4: SDS depletion efficiency following protein precipitation. The percentage of SDS removed is shown from acetone precipitation with the conventional protocol (overnight at -20 °C), the rapid protocol (2 min incubation at room temperature), or by chloroform/methanol/water (CMW) precipitation of an S. cerevisiae lysate, both in conventional (vial) and cartridge format. These samples initially contained 0.5% SDS (5,000 ppm), inferring >99.8% SDS removal is required for optimal MS analysis. Residual SDS is quantified by methylene blue active substances (MBAS) assay. Error bars represent the standard deviation from technical replicates (n = 3). Please click here to view a larger version of this figure.
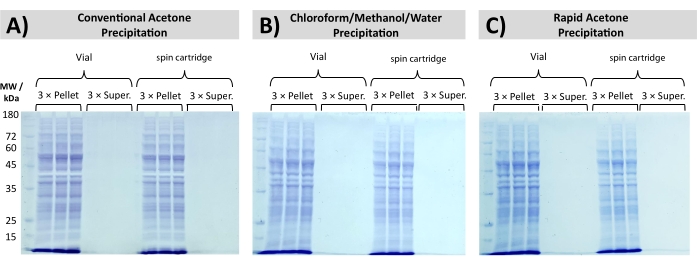
Figure 5: Total proteome recovery through precipitation. SDS PAGE shows the recovery of S. cerevisiae total protein lysate, precipitated by (A) conventional acetone precipitation, (B) chloroform/methanol/water precipitation, and (C) rapid acetone precipitation. Protein bands are exclusively observed in the pellet fraction, with no visible bands in the supernatant (Super.). Please click here to view a larger version of this figure.
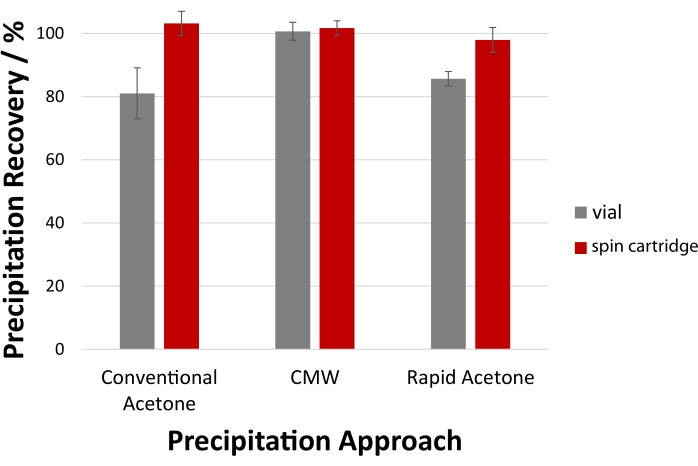
Figure 6: Superior protein recovery within a filtration cartridge. For precipitation of the S. cerevisiae total protein lysate, the disposable spin cartridge facilitates quantitative recovery with acetone and CMW precipitation. High recovery is also possible with vial-based precipitation, though careful sample manipulation and pipetting are required. LC-UV assessed protein recovery following resolubilization of the pellet with cold formic acid are shown here. Error bars represent the standard deviation from technical replicates (n = 3). Please click here to view a larger version of this figure.
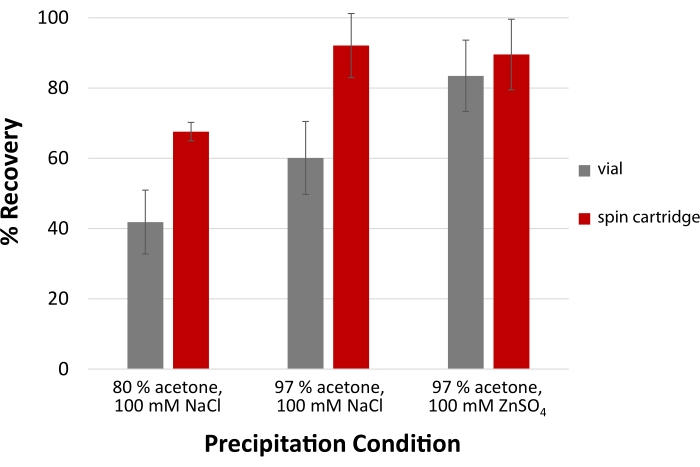
Figure 7: High precipitation yields for low molecular weight peptides. A modified acetone precipitation protocol for peptides and proteins ≤5 kDa involves coupling 100 mM of ZnSO4 with 97% acetone to achieve the highest yields. Precipitation facilitated by a disposable filtration cartridge demonstrates improved recovery compared to conventional vial-based precipitation across all three precipitation conditions. Error bars represent the standard deviation from technical replicates (n = 3). Please click here to view a larger version of this figure.
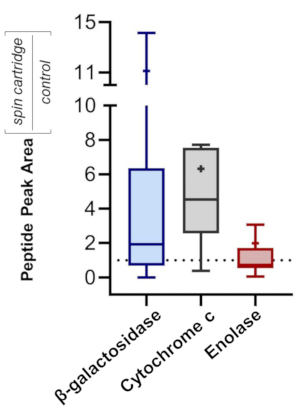
Figure 8: Higher recovery of standard proteins in SDS-based workflow. Tukey Box-and-Whisker plots30 of relative MS signal intensity for peptides recovered from SDS-containing proteins processed in a disposable filtration cartridge relative to a control sample (no SDS, no precipitation). The proteins employed span a wide concentration dynamic range-β-galactosidase:cytochrome c:enolase = 10,000:10:1. Each quartile within the boxes contains 25% of the distribution, while error bars encompass 95% of the distribution. Mean is indicated by "+" and median by a horizontal line. Please click here to view a larger version of this figure.
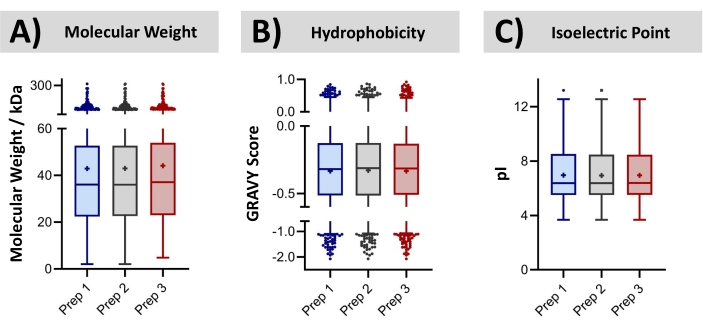
Figure 9: Identified protein distributions from technical replicates. Tukey Box-and-Whisker plots characterize (A) the molecular weight, (B) hydrophobicity, and (C) isoelectric point of proteins identified by bottom-up LC-MS/MS following triplicate preparations of a bovine liver lysate in a two-stage filtration and extraction cartridge. There was no statistical difference in these characteristics by two-way ANOVA (p < 0.05). Each quartile within the boxes contains 25% of the distribution, while error bars encompass 95% of the distribution. Please click here to view a larger version of this figure.
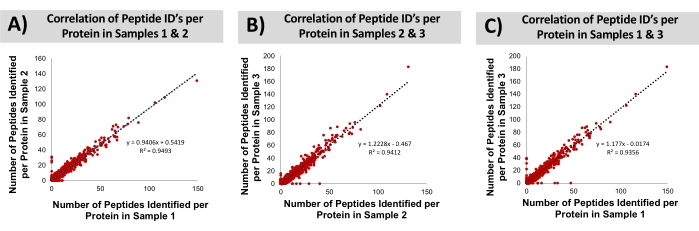
Figure 10: Correlation of peptide IDs per protein through the SDS-based preparation workflow across preparative replicates. Analysis of bottom-up proteome reproducibility across (A) samples 1 and 2, (B) samples 2 and 3, and (C) samples 1 and 3 based on the number of peptide MS identifications per protein. Please click here to view a larger version of this figure.
Discussion
Optimal MS characterization is achieved when residual SDS is depleted below 10 ppm. While alternative approaches, such as FASP and on-bead digestion, offer quantitative SDS depletion with variable recovery31,32,33, the primary objective of precipitation is to maximize purity and yield simultaneously. This depends on effectively isolating the supernatant (containing the SDS) without disturbing the protein pellet. With vial-based precipitation, once the bulk of the supernatant is removed by pipetting, it is increasingly likely that some of the aggregated pellets are accidentally lost. For this reason, it is essential to leave behind a more significant fraction of the residual solvent (~20 µL) and to add a washing step34. The washing step dilutes and removes the residual solvent from the vial. Particularly with CMW, it is unnecessary to vortex the sample vial once the pellet has formed. Disrupting the pellet through vigorous agitation has the unwanted effect of increasing the likelihood of loss from accidental pipetting. If vortexing is included (as recommended by previous protocols)35, the potential exists for portions of the CMW pellet to adhere to the underside of the vial cap; once centrifuged, the pellet remains fixed on the vial cap and can result in ~50% loss.
Rapid precipitation can be performed with high recovery of dilute proteome samples, ideally between 0.01-2 mg/mL, or a corresponding protein mass between 1-200 µg. However, quantitative and reproducible recoveries starting from below 0.01 mg/mL protein may benefit from longer precipitation times ranging from 10 min to 1 h, demonstrating throughput limitations of the precipitation workflow. Surprisingly, more concentrated samples (10 mg/mL) show a statistical reduction in yield, presumably from accidental pipetting losses. Assuming >10 µg protein, a visible pellet should be observed on the side of the vial (Figure 1B). Smaller quantities, down to 1 µg, are challenging to see. This challenges the capacity to pipette the supernatant without disrupting the protein pellet. The vial can be inverted (slowly) with acetone to separate the solvent from the pellet. For CMW, the pellet does not reliably adhere sufficiently to the vial, thereby favoring pipetting over decanting of the supernatant. For vial-based precipitation, working with the smallest possible microcentrifuge tube is recommended to facilitate the intended sample and solvent volumes. Precipitation in the disposable filtration cartridge employed in this work provides a maximum volume capacity of 500 µL, enabling protein precipitation with 80% acetone on sample volumes up to 100 µL. Sample, salt, and solvent volumes can be adjusted accordingly if the recommended concentrations are maintained.
The purity of the protein pellet recovered from organic solvent-based precipitation is limited by the complexity of the sample matrix, buffer components, and precipitation conditions. For example, specific buffer components such as glycine (used for SDS PAGE separations) have been shown to co-precipitate with protein using 80% acetone. However, glycine remains soluble through CMW precipitation. Acetone has been reported to precipitate DNA fragments36,37, potentially adding undesired background impurities to the recovered pellet. Precipitation of low molecular weight proteins and peptides requires an elevated level of organic solvent and a specific salt type to maximize yield. While several salts have been explored, ZnSO4 provides consistently high products. This salt will precipitate in 97% acetone in the absence of protein. Thus, the resulting protein pellet contains a high concentration of salt. It is noted that employing 90% acetone by volume will also achieve high peptide yields, though a statistically significant drop (~5%) in recovery is expected. However, this allows processing a more significant sample volume (up to 180 µL, with 20 µL of 1 M ZnSO4) in each 2 mL vial. Beyond matrix impurities co-precipitating with the protein pellet, it must be stated that solvent precipitation inherently causes denaturation of the sample38,39. Therefore, this protocol is not applicable for the preparations of functional proteins or native MS workflows. Acetone has also been reported to cause covalent protein modifications at glycine residues40 and induce a +98 u mass shift, speculated to be a byproduct of aldol condensation acetone41.
When employing a filtration cartridge for protein precipitation, isolation of the protein pellet relies on the retention of aggregates above a PTFE membrane filter. The porosity of this membrane exceeds that of a molecular weight cutoff filter (as seen in FASP), permitting protein isolation with reduced spin times. Rapid solution transfer at low spin speeds relies on proper wetting of the PTFE membrane; organic solvents readily flow through, though a dry PTFE filter impedes aqueous solvents. If the filtration cartridge appears to be clogged, the membrane should be re-wetted by directly applying a small volume of organic solvent (e.g., isopropanol) to the filter. Depending on the size of the protein pellet and the volume of sample employed, additional centrifugation or spins at higher speeds (up to 3,000 x g) may be required to ensure all solvent has passed through the filtration cartridge.
Protein recovery from precipitation with optimal conditions is ultimately limited by the challenge of pellet re-solubilization, with few solvent options being compatible with downstream processing and LC-MS. Additionally, several precipitation conditions such as long exposures to low temperatures and over-drying a tightly packed pellet contribute to re-solubilization challenges40. It is noted that CMW protein pellets are generally less soluble than acetone pellets. Maximized re-solubilization efficiency of precipitated protein by 80% cold formic acid (step 6.2.2) has previously been reported42; the cold temperature prevents protein modification, which otherwise occurs in concentrated formic acid43,44. Diluting the acid concentration also slows the modification reaction. Formic acid is recommended for top-down MS approaches or before enzymatic digestion with pepsin. Employing this solvent demands little physical treatment; the addition of only 5 µL (enough to cover the protein pellet) may be sufficient when combined with vortexing, brief sonication, or repeat pipetting. Similarly, for samples intended to be analyzed by SDS PAGE, re-dissolving in SDS-containing Laemmli buffer is highly effective, when combined with modest mixing of the sample prior to heating. However, these solvents are both incompatible with trypsin. Resolubilization with 8 M urea is recommended prior to trypsin digestion, ensuring that the urea has been freshly prepared (same day). A minimum volume of 50 µL buffer is recommended for protein re-solubilization within the filtration cartridge to maximize contact between the chaotropic solvent and pellet, as well as to aid dissolution during sonication, repeat pipetting and/or vortexing. Alternative approaches exploit trypsin to re-solubilize the protein, meaning the protein need not be fully re-dissolved prior to enzyme addition. However, this approach can bias digestion, favoring the more soluble species while hydrophobic proteins experience shorter digestion time45. Addition of 8 M urea, together with basic buffers such as Tris or ammonium bicarbonate, demands a post-digestion sample clean-up step. For such sample additives, reversed phase column clean-up is ideal. The disposable filtration cartridge employed in this study is supplemented with an interchangeable reversed phase SPE cartridge. This cartridge is also ideally suited for solvent exchange, in the case of the formic acid resolubilization protocol. It is important to note that any solid phase extraction approach is associated with inherent loss in sample recovery. Therefore, the user should weigh the benefits of recovery and the additional purification for their experiment.
It is anticipated that these protocols will enable proteomics researchers to streamline their detergent-based workflows, capitalizing on SDS for proteome extraction. Preparative strategies that facilitate consistent recovery of the complete proteome are critical. A two-stage spin cartridge simplifies the opportunity for rapid, robust, and reproducible proteome sample isolation. Such an approach would be amenable to applications requiring rigorous analysis without sacrificing sample throughputs, such as clinical settings or large-scale research initiatives46. Future applications of these approaches may include biomarker discovery, detection, accurate quantitation, and drug and drug target discovery.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work was funded by the Natural Sciences and Engineering Research Council of Canada. The authors thank Bioinformatics Solutions Inc. (Waterloo, Canada) and SPARC BioCentre (Molecular Analysis) at the Hospital for Sick Children (Toronto, Canada) for their contributions to the acquisition of MS data.
Materials
| Acetone | Fisher Scientific | AC177170010 | ≤0.002 % aldehyde |
| Acetonitrile | Fisher Scientific | A998-4 | HPLC grade |
| Ammonium Bicarbonate | Millipore Sigma | A6141-1KG | solid |
| Beta mercaptoethanol | Millipore Sigma | M3148-25ML | Molecular biology grade |
| Bromophenol blue | Millipore Sigma | B8026-5G | Bromophenol blue sodium salt |
| Chloroform | Fisher Scientific | C298-400 | Chloroform |
| Formic Acid | Honeywell | 56302 | Eluent additive for LC-MS |
| Fusion Lumos Mass Spectrometer | ThermoFisher Scientific | for analysis of standard protein mixture | |
| Glycerol | Millipore Sigma | 356352-1L-M | For molecular biology, > 99% |
| Isopropanol | Fisher Scientific | A4641 | HPLC grade |
| Methanol | Fisher Scientific | A452SK-4 | HPLC grade |
| Microcentrifuge | Fisher Scientific | 75-400-102 | up to 21,000 xg |
| Microcentrifuge Tube (1.5 mL) | Fisher Scientific | 05-408-130 | tapered bottom |
| Microcentrifuge Tube (2 mL) | Fisher Scientific | 02-681-321 | rounded bottom |
| Micropipette Tips (0.1-10 μL) | Fisher Scientific | 21-197-28 | Universal pipet tip, non-sterile |
| Micropipette Tips (1-200 μL) | Fisher Scientific | 07-200-302 | Universal pipet tip, non-sterile |
| Micropipette Tips (200-1000 μL) | Fisher Scientific | 07-200-303 | Universal pipet tip, non-sterile |
| Micropipettes | Fisher Scientific | 13-710-903 | Micropipet Trio pack |
| Pepsin | Millipore Sigma | P0525000 | Lyophilized powder, >3200 units/ mg |
| ProTrap XG | Proteoform Scientific | PXG-0002 | 50 complete units per box |
| Sodium Chloride | Millipore Sigma | S9888-1KG | ACS reagent, >99 % |
| Sodium Dodecyl Sulfate | ThermoFisher Scientific | 28312 | powdered solid |
| timsTOF Pro Mass Spectrometer | Bruker | for analysis of liver proteome extract | |
| Trifluoroacetic Acid | ThermoFisher Scientific | L06374.AP | 99% |
| Tris | Fisher Scientific | BP152-500 | Molecular biology grade |
| Trypsin | Millipore Sigma | 9002-07-7 | From bovine pancreas, TPCK-treated |
| Urea | Bio-Rad | 1610731 | solid |
| Water (deionized) | Sartorius Arium Mini Water Purification System | 76307-662 | Type 1 ultrapure (18.2 MΩ cm) |
| Zinc Sulfate | Millipore Sigma | 307491-100G | solid |
Referencias
- Kilpatrick, L. E., Kilpatrick, E. L. Optimizing high-resolution mass spectrometry for the identification of low-abundance post-translational modifications of intact proteins. Journal of Proteome Research. 16 (9), 3255-3265 (2017).
- Scheffler, K., Viner, R., Damoc, E. High resolution top-down experimental strategies on the Orbitrap platform. Journal of Proteomics. 175, 42-55 (2018).
- Quaranta, A., et al. N -Glycosylation profiling of intact target proteins by high-resolution mass spectrometry (MS) and glycan analysis using ion mobility-MS/MS. Analyst. 145 (5), 1737-1748 (2020).
- Van Der Burgt, Y. E. M., et al. Structural analysis of monoclonal antibodies by ultrahigh resolution MALDI in-source decay FT-ICR mass spectrometry. Analytical Chemistry. 91 (3), 2079-2085 (2019).
- Anderson, L. C., et al. Identification and characterization of human proteoforms by top-down LC-21 Tesla FT-ICR mass spectrometry. Journal of Proteome Research. 16 (2), 1087-1096 (2017).
- Nickerson, J. L., et al. Recent advances in top-down proteome sample processing ahead of MS analysis. Mass Spectrometry Reviews. , (2021).
- Kelly, R. T. Single-cell Proteomics: Progress and Prospects. Molecular and Cellular Proteomics. 19 (11), 1739-1748 (2020).
- Alexovič, M., Sabo, J., Longuespée, R. Microproteomic sample preparation. Proteomics. 21 (9), 2000318 (2021).
- Shishkova, E., Coon, J. J. Rapid preparation of human blood plasma for bottom-up proteomics analysis. STAR Protocols. 2 (4), 100856 (2021).
- Duong, V. A., Park, J. M., Lee, H. Review of three-dimensional liquid chromatography platforms for bottom-up proteomics. International Journal of Molecular Sciences. 21 (4), 1524 (2020).
- Gan, G., et al. SCASP: A simple and robust SDS-aided sample preparation method for proteomic research. Molecular and Cellular Proteomics. 20, 100051 (2021).
- Kachuk, C., Doucette, A. A. The benefits (and misfortunes) of SDS in top-down proteomics. Journal of Proteomics. 175, 75-86 (2018).
- Rundlett, K. L., Armstrong, D. W. Mechanism of signal suppression by anionic surfactants in capillary electrophoresis-electrospray ionization mass spectrometry. Analytical Chemistry. 68 (19), 3493-3497 (1996).
- Wis, J. R., Zougman, A., Nagaraj, N., Mann, M. Universal sample preparation method for proteome analysis. Nature Methods. 6 (5), 359-362 (2009).
- Ni, M., et al. Modified filter-aided sample preparation (FASP) method increases peptide and protein identifications for shotgun proteomics. Rapid Communications in Mass Spectrometry. 31 (2), 171-178 (2017).
- Zhao, Q., et al. imFASP: An integrated approach combining in-situ filter-aided sample pretreatment with microwave-assisted protein digestion for fast and efficient proteome sample preparation. Analytica Chimica Acta. 912, 58-64 (2016).
- Kanshin, E., et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Molecular Systems Biology. 10 (10), 757 (2014).
- Hughes, C. S., et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nature Protocols. 14 (1), 68-85 (2019).
- Dagley, L. F., Infusini, G., Larsen, R. H., Sandow, J. J., Webb, A. I. Universal solid-phase protein preparation (USP3) for bottom-up and top-down proteomics. Journal of Proteome Research. 18 (7), 2915-2924 (2019).
- Hengel, S. M., et al. Evaluation of SDS depletion using an affinity spin column and IMS-MS detection. Proteomics. 12 (21), 3138-3142 (2012).
- Zougman, A., Selby, P. J., Banks, R. E. Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. PROTEOMICS. 14 (9), 1006-1010 (2014).
- Thongboonkerd, V., McLeish, K. R., Arthur, J. M., Klein, J. B. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney International. 62 (4), 1461-1469 (2002).
- Klont, F., et al. Assessment of sample preparation bias in mass spectrometry-based proteomics. Analytical Chemistry. 90 (8), 5405-5413 (2018).
- Crowell, A. M. J., Wall, M. J., Doucette, A. A. Maximizing recovery of water-soluble proteins through acetone precipitation. Analytica Chimica Acta. 796, 48-54 (2013).
- Nickerson, J. L., Doucette, A. A. Rapid and quantitative protein precipitation for proteome analysis by mass spectrometry. Journal of Proteome Research. 19 (5), 2035-2042 (2020).
- Baghalabadi, V., Doucette, A. A. Mass spectrometry profiling of low molecular weight proteins and peptides isolated by acetone precipitation. Analytica Chimica Acta. 1138, 38-48 (2020).
- Crowell, A. M. J., MacLellan, D. L., Doucette, A. A. A two-stage spin cartridge for integrated protein precipitation, digestion and SDS removal in a comparative bottom-up proteomics workflow. Journal of Proteomics. 118, 140-150 (2015).
- Wessel, D., Flügge, U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Analytical Biochemistry. 138 (1), 141-143 (1984).
- Arand, M., Friedberg, T., Oesch, F. Colorimetric quantitation of trace amounts of sodium lauryl sulfate in the presence of nucleic acids and proteins. Analytical Biochemistry. 207 (1), 73-75 (1992).
- Tukey, J. W. Exploratory data analysis. Biometrics. 33, 768 (1977).
- Ludwig, K. R., Schroll, M. M., Hummon, A. B. Comparison of in-solution, FASP, and S-Trap based digestion methods for bottom-up proteomic studies. Journal of Proteome Research. 17 (7), 2480-2490 (2018).
- Sielaff, M., et al. Evaluation of FASP, SP3, and iST protocols for proteomic sample preparation in the low microgram range. Journal of Proteome Research. 16 (11), 4060-4072 (2017).
- Supasri, K. M., et al. Evaluation of filter, paramagnetic, and STAGETips aided workflows for proteome profiling of symbiodiniaceae. Processes. 9 (6), 983 (2021).
- Botelho, D., et al. Top-down and bottom-up proteomics of SDS-containing solutions following mass-based separation. Journal of Proteome Research. 9 (6), 2863-2870 (2010).
- Fic, E., Kedracka-Krok, S., Jankowska, U., Pirog, A., Dziedzicka-Wasylewska, M. Comparison of protein precipitation methods for various rat brain structures prior to proteomic analysis. Electrophoresis. 31 (21), 3573-3579 (2010).
- Antonioli, P., Bachi, A., Fasoli, E., Righetti, P. G. Efficient removal of DNA from proteomic samples prior to two-dimensional map analysis. Journal of Chromatography A. 1216 (17), 3606-3612 (2009).
- Bryzgunova, O., et al. A reliable method to concentrate circulating DNA. Analytical Biochemistry. 408 (2), 354-356 (2011).
- Asakura, T., Adachi, K., Schwartz, E. Stabilizing effect of various organic solvents on protein. Journal of Biological Chemistry. 253 (18), 6423-6425 (1978).
- Loo, J. A., Loo, R. R. O., Udseth, H. R., Edmonds, C. G., Smith, R. D. Solvent-induced conformational changes of polypeptides probed by electrospray-ionization mass spectrometry. Rapid Communications in Mass Spectrometry. 5 (3), 101-105 (1991).
- Simpson, D. M., Beynon, R. J. Acetone precipitation of proteins and the modification of peptides. Journal of Proteome Research. 9 (1), 444-450 (2010).
- Güray, M. Z., Zheng, S., Doucette, A. A. Mass spectrometry of intact proteins reveals +98 u chemical artifacts following precipitation in acetone. Journal of Proteome Research. 16 (2), 889-897 (2017).
- Doucette, A. A., Vieira, D. B., Orton, D. J., Wall, M. J. Resolubilization of precipitated intact membrane proteins with cold formic acid for analysis by mass spectrometry. Journal of Proteome Research. 13 (12), 6001-6012 (2014).
- Beavis, R. C., Chait, B. T. Rapid, sensitive analysis of protein mixtures by mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 87 (17), 6873-6877 (1992).
- Klunk, W. E., Pettegrew, J. W. Alzheimer’s β-Amyloid protein is covalently modified when dissolved in formic acid. Journal of Neurochemistry. 54 (6), 2050-2056 (1990).
- Vuckovic, D., Dagley, L. F., Purcell, A. W., Emili, A. Membrane proteomics by high performance liquid chromatography-tandem mass spectrometry: Analytical approaches and challenges. Proteomics. 13 (3-4), 404-423 (2013).
- Smith, L. M., et al. The human proteoform project: Defining the human proteome. Science Advances. 7 (46), (2021).

