Quantifying Cytoskeleton Dynamics Using Differential Dynamic Microscopy
Summary
Differential dynamic microscopy (DDM) combines features of dynamic light scattering and microscopy. Here, the process of using DDM to characterize reconstituted cytoskeleton networks by quantifying the subdiffusive and caged dynamics of particles in vimentin networks and the ballistic motion of active myosin-driven actin-microtubule composites is presented.
Abstract
Cells can crawl, self-heal, and tune their stiffness due to their remarkably dynamic cytoskeleton. As such, reconstituting networks of cytoskeletal biopolymers may lead to a host of active and adaptable materials. However, engineering such materials with precisely tuned properties requires measuring how the dynamics depend on the network composition and synthesis methods. Quantifying such dynamics is challenged by variations across the time, space, and formulation space of composite networks. The protocol here describes how the Fourier analysis technique, differential dynamic microscopy (DDM), can quantify the dynamics of biopolymer networks and is particularly well suited for studies of cytoskeleton networks. DDM works on time sequences of images acquired using a range of microscopy modalities, including laser-scanning confocal, widefield fluorescence, and brightfield imaging. From such image sequences, one can extract characteristic decorrelation times of density fluctuations across a span of wave vectors. A user-friendly, open-source Python package to perform DDM analysis is also developed. With this package, one can measure the dynamics of labeled cytoskeleton components or of embedded tracer particles, as demonstrated here with data of intermediate filament (vimentin) networks and active actin-microtubule networks. Users with no prior programming or image processing experience will be able to perform DDM using this software package and associated documentation.
Introduction
The cytoskeleton is a network of protein filaments that spans across the cytoplasm of eukaryotic cells, connecting the cell surface to the nucleus. It has unique material properties, providing mechanical protection against large and repeated mechanical loads, yet also driving dynamic cell shape changes1. Reconstituted cytoskeleton networks can give rise to a range of interesting dynamic behaviors, from the caging of embedded particles to ballistic motion driven by molecular motors2,3,4,5,6,7,8,9,10,11. Methods to analyze the dynamics of such networks include tracking the motion of embedded tracer microspheres6,7,12,13,14, image analysis to track the size of protein-dense clusters over time8, dynamic light scattering15, particle image velocimetry4,16,17,18,19, computing the power spectral density of images over time19, and kymograph analysis20. As more studies on reconstituted cytoskeleton networks are conducted, whether to understand cellular mechanics or active matter, robust, unbiased, and reproducible methods for characterizing the dynamics are increasingly necessary. Differential dynamic microscopy (DDM)21,22, a relatively new technique that has been used to study cytoskeleton dynamics, is one such technique that efficiently quantifies dynamics with few user-defined parameters. With the software package described here, researchers with little experience in programming or image analysis will be able to leverage DDM for their own work.
DDM is an image analysis technique to extract a sample's dynamics. Like particle tracking or particle image velocimetry, DDM requires a time series of images (often thousands of images), typically recorded with a microscope. Unlike particle tracking, individual features or tracer beads do not need to be localized (or even be localizable) in the image. Unlike both particle tracking and particle image velocimetry, one recovers the ensemble dynamics with DDM with relatively few user-specified parameters. With DDM, images are analyzed in Fourier space to determine the decay time of density fluctuations over a range of wavenumbers, q, where q = 2πu, and u is the magnitude of the spatial frequencies,  . One obtains scattering-like information but with real-space images acquired on a microscope21,22,23. Therefore, one can take advantage of the various contrast-generating methods of microscopy, such as widefield fluorescence22,24, confocal fluorescence25, polarized26, dark-field27, or light-sheet fluorescence28 microscopies. Furthermore, images used for DDM analysis may be used for particle tracking or particle image velocimetry to provide complementary information.
. One obtains scattering-like information but with real-space images acquired on a microscope21,22,23. Therefore, one can take advantage of the various contrast-generating methods of microscopy, such as widefield fluorescence22,24, confocal fluorescence25, polarized26, dark-field27, or light-sheet fluorescence28 microscopies. Furthermore, images used for DDM analysis may be used for particle tracking or particle image velocimetry to provide complementary information.
This combination of features from dynamic light scattering and optical microscopy makes DDM a powerful and versatile technique. Since its first description by Cerbino and Trappe in 200821, where DDM was demonstrated to measure the diffusion of 73 nm colloidal particles, DDM has been used to measure flowing colloids29, colloidal aggregation30,31, the viscoelasticity of nematic liquid crystals26, the dynamics of colloidal gels32, coarsening foams33, nanoparticles in confined environments34,35,36,37, bacterial motility38,39,40,41, the diffusion of weakly scattering protein clusters42, capillary waves at fluid interfaces43, and other systems. Those looking for a more complete listing of publications employing DDM can refer to thorough review papers on the subject22,23,44,45.
DDM has also been used to investigate the dynamics of biological networks. Drechsler et al. used DDM to measure the dynamics of actin in living Drosophila oocytes46. Burla et al. quantified the dynamics of tracer particles in networks of hyaluronan and hyaluronan-collagen composites47. Several uses of DDM to study the dynamics of tracer particles in reconstituted cytoskeleton networks9,10, the transport of DNA molecules in such networks48,49, and the dynamics of active reconstituted networks have also been documented11,50,51. An advantage of DDM in measuring the dynamics in such systems is that individual particles or molecules do not need to be localized and tracked. So, for example, the dynamics of DNA molecules in crowded environments can be measured with DDM despite the difficulty in tracking such small and non-spherical molecules. Furthermore, with fluorescence microscopy, one can use multi-color labeling to selectively measure the dynamics of individual constituents in a complex composite.
To perform DDM, a sequence of images is taken over time, I(x,y,t). For a given lag time, Δt, all (or a subset of) pairs of images separated by that lag time are found. The squared Fourier transform of the difference of each pair,

is computed and averaged together. This quantity,  , is radially averaged, provided that the dynamics are isotropic. This yields the DDM matrix (also referred to as the image structure function),
, is radially averaged, provided that the dynamics are isotropic. This yields the DDM matrix (also referred to as the image structure function),  . This process is shown graphically in Figure 1. To determine the sample's dynamics from this DDM matrix, the DDM matrix is assumed to take the form
. This process is shown graphically in Figure 1. To determine the sample's dynamics from this DDM matrix, the DDM matrix is assumed to take the form

where A is the amplitude, which depends on the details of the microscope and the structure of the sample, B is the background, which depends on the noise in the images, and f (q, Δt) is the intermediate scattering function (ISF), which contains information about the dynamics21,22. In simple cases,

where τ is a characteristic decay or decorrelation time. Such an ISF has been used in several studies employing DDM on ergodic systems like dilute colloidal suspensions21,24,27,37,40,52. However, other forms of the ISF can be used to model various types of dynamics. For example, one may use a cumulant expansion to model the ISF for polydisperse samples as

where μ is a measure of the polydispersity42,53; if density fluctuations decay by two separate modes one may use an ISF like
 26, 54, 55, 56, 57;
26, 54, 55, 56, 57;
other ISFs can be used for swimming micro-organisms or other active particles38,39,40,41,58,59.

Figure 1: Overview of DDM analysis. From the time series of images, the Fourier transform of image differences is computed to calculate the DDM matrix. The DDM matrix can be fit to a model to determine the time scale of density fluctuations across a range of q values. Please click here to view a larger version of this figure.
Here, the use of a DDM analysis software package developed in Python, PyDDM, is described. This software package builds on the work done by our research labs and other published studies over the last several years. Primary motivators for creating this software package include the need for (1) keeping track of and storing metadata and parameters used in the analysis; (2) thorough documentation with detailed examples of analysis from start to finish; and (3) an easy way to employ different (or create new) mathematical models for fitting the data (e.g., adding ISF models, such as ones recently developed for active filaments60, would be straightforward). Other software packages for DDM analysis also exist, though not all are well documented and written in an open-source programming language. For example, there is C++ code with computing on GPUs (https://github.com/peterlu/ConDDM)25, C++ code that uses Fourier transforms in time to speed up calculations (https://github.com/giovanni-cerchiari/diffmicro)61, MATLAB and Python versions (https://github.com/MathieuLeocmach/DDM)40, MATLAB code (https://sites.engineering.ucsb.edu/~helgeson/ddm.html)27, and MATLAB code with uncertainty quantification (https://github.com/UncertaintyQuantification/DDM-UQ)62. As this PyDDM package is well documented and provides a lot of flexibility in how the DDM matrix is calculated and analyzed, it can hopefully be useful to researchers looking to implement DDM regardless of their background in programming or image analysis.
The protocol shows how this software package can be used to quantify the dynamics of in vitro reconstituted cytoskeleton networks. This is done by using two distinct sets of imaging data: (1) images of submicron tracer particles embedded in a vimentin network taken with brightfield microscopy and (2) images of fluorescently labeled actin and microtubule filaments in an entangled composite network with myosin-driven activity taken with laser-scanning confocal microscopy. The analyses of these two datasets highlight notable strengths of DDM, including its ability to analyze images taken with a variety of imaging modalities (e.g., brightfield or confocal fluorescence), to extract dynamics from either embedded tracers or from labeled filaments, and to quantify a variety of dynamics (e.g., subdiffusive and constrained or ballistic).
Protocol
NOTE: A Jupyter Notebook file containing the code to go along with each step in the following protocol can be found on the following GitHub repository, https://github.com/rmcgorty/PyDDM/tree/main/Examples. A PDF of that file is included in Supplementary File 1. Additionally, a walkthrough of the code and documentation of each function and class can be found on the website, https://rmcgorty.github.io/PyDDM/.
1. Software installation
- To follow along with the example DDM analysis files, install Jupyter Notebook for running the code. Install other required common Python packages, including NumPy and Matplotlib as well. These packages all come bundled with the Anaconda distribution (see https://www.anaconda.com/products/individual).
- Install the Python package xarray63. This package is necessary for organizing and storing metadata and analysis parameters. If using the Anaconda distribution, install xarray (along with its recommended dependencies) using the command:
conda install -c conda-forge xarray dask netCDF4 bottleneck - Install the PyYAML package using the command:
conda install -c anaconda yaml
This package is necessary for reading metadata about the images to analyze and the parameters set by the user for analysis and fitting. - Install the PyDDM package, by downloading from the GitHub repository or using the git command:
git clone https://github.com/rmcgorty/PyDDM.git
2. Planning the imaging sessions
- Choose the optimal available imaging modality and optical settings. As mentioned, DDM can be used with a number of microscopy methods.
- To assist in planning the appropriate objective lens and image size to use, determine the range of wavenumbers, q, that will be probed based on the pixel size and total image size. Confirm that the choice of magnification and field of view are optimal for the experiment, based on these calculations. For the images analyzed here, a 60x 1.4 NA objective and an image size of 256 x 256 pixels with a pixel size of 0.83 µm were used for the active actin-microtubule composite network. For the images of beads embedded in a vimentin network, a 100x 1.4 NA objective and an image size of 512 x 512 pixels with a pixel size of 0.13 µm were used.
NOTE: The minimum q is set by 2π/NΔx, where the image size (assumed to be square) is N × N pixels with a pixel size of Δx. The maximum q is the minimum of π/Δx and 2π NA/λ, where NA is the imaging objective's numerical aperture, and λ is the wavelength of light (for brightfield imaging, one can replace NA with (NAobjective + NAcondenser)/2). - Next, consider the range of timescales to investigate. Typically, DDM analysis is done on sequences of at least 1000 frames.
- To determine the appropriate frame rate, consider the expected time it will take for features in the sample to move a distance on the order of the minimum resolvable length scale (corresponding to the maximum q).
- In considering the upper limit of the range of timescales probed, recognize that, typically, the power spectrum of hundreds of image differences of a given lag time Δt are averaged together to provide sufficient statistics to reduce noise. Hence, acquire image sequences longer than the maximum timescale probed.
NOTE: If an expected diffusion coefficient, D, or velocity, v, is known, then one can estimate expected characteristic decay times using τ = 1/Dq2 or τ = 1/vq along with the range of q, which was determined based on the field of view and pixel size. The range of expected τ values over the accessible q-range can help guide the choice of frame rate and the number of frames to acquire.
3. Sample preparation and image acquisition
NOTE: For details of the sample preparation and imaging settings used for the data presented in the representative results section, see previous publications from the authors11,51,64 and Supplementary File 2.
- Based on the consideration of the time and length scales to probe, acquire image sequences of, ideally, over 1000 frames.
NOTE: The code will analyze square images or square regions of interest within the image, so adjust the frame size accordingly. - Save image sequences as a three-dimensional greyscale TIFF stack. Alternatively, the format used by Nikon Instruments systems, ND2 format, can be read by the installed package. If images are saved in some different format, use ImageJ or another imaging processing program to convert the images to a TIFF stack.
NOTE: If using ND2 files, the package nd2reader from https://github.com/Open-Science-Tools/nd2reader must be installed.
4. Parameter setup
- Make a copy of the parameter file example_parameter_file.yml provided in the PyDDM code repository under the examples folder. Open this YAML file with a text editor like NotePad++ or the text editor in JupyterLab. See Supplementary File 2 for an example YAML parameters file used in the analysis of data presented in the representative results section.
- In the copied YAML file, provide the data directory and file name corresponding to the image sequence to be analyzed. Under the metadata section, provide the pixel size and frame rate.
- Under the Analysis_parameters section, provide details for how the DDM matrix should be calculated. Some parameters here are optional.
- At a minimum, provide values for the parameters number_lag_times and last_lag_time. These correspond to the number of different lag times for which to calculate the DDM matrix and the longest lag time (in frames) to use, respectively. For the data of tracer beads in vimentin networks used here, the parameters number_lag_times and last_lag_time were 60 and 1000, respectively. The code will compute the DDM matrix for lag times from 1 frame (or some other minimum lag time if the optional parameter first_lag_time is specified) to the last_lag_time with logarithmic spacing.
NOTE: If M frames were acquired, one could calculate the DDM matrix for a lag time as large as M-1. However, with poor statistics at such a large lag time, the data is likely to be noisy. The longest lag time for which to calculate the DDM matrix will depend on the details of the data, but we suggest trying around one-third of the total image series duration.
- At a minimum, provide values for the parameters number_lag_times and last_lag_time. These correspond to the number of different lag times for which to calculate the DDM matrix and the longest lag time (in frames) to use, respectively. For the data of tracer beads in vimentin networks used here, the parameters number_lag_times and last_lag_time were 60 and 1000, respectively. The code will compute the DDM matrix for lag times from 1 frame (or some other minimum lag time if the optional parameter first_lag_time is specified) to the last_lag_time with logarithmic spacing.
- Provide details for how the DDM matrix or the intermediate scattering function (ISF) should be fit in the Fitting_parameters section. Give the name of the model under the model parameter. Provide the initial guess, lower bound, and upper bound for each of the fitting parameters in the chosen model.
NOTE: To display a list of the possible fitting models, run the function print_fitting_models. The models can also be found in the online documentation on the PyDDM website.
5. Calculating the DDM matrix
- Initialize an instance of the DDM_Analysis class. To do so, provide the metadata and analysis parameters discussed above by passing the filename, with the full file path included, of the YAML file to DDM_Analysis. Alternatively, pass the metadata and parameters as a Python dictionary data structure.
- Run the function calculate_DDM_matrix to calculate the DDM matrix. This calculation may take several minutes or longer depending on the frame size and the number of lag times. See Figure 2 for typical run times.
- Inspect the returned data, which will be in a data structure from the xarray package known as a Dataset. This data structure is stored under the attribute ddm_dataset.
NOTE: Not only the DDM matrix but also associated variables and metadata will be stored in this data structure. It will also be saved to disk in a Network Common Data Form (netCDF) format. - Inspect the plots and figures, which will be generated and displayed. These figures are also saved as a PDF file in the data directory.
- See that one of the generated plots shows the ensemble-averaged squared modulus of the Fourier-transformed images,
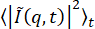 as a function of q. By default, the code uses this to estimate the background parameter B. Estimate the background from
as a function of q. By default, the code uses this to estimate the background parameter B. Estimate the background from 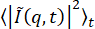 by assuming that, in the limit of large q, it will approach B/2, where B is the background.
by assuming that, in the limit of large q, it will approach B/2, where B is the background. - If
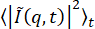 is not reaching a plateau at large q, then use another method for estimating B. To accomplish this, set the parameter background_method in either the YAML file or as an optional keyword argument to the function calculate_DDM_matrix. More details about the methods for estimating B are presented in the representative results section.
is not reaching a plateau at large q, then use another method for estimating B. To accomplish this, set the parameter background_method in either the YAML file or as an optional keyword argument to the function calculate_DDM_matrix. More details about the methods for estimating B are presented in the representative results section.
- See that one of the generated plots shows the ensemble-averaged squared modulus of the Fourier-transformed images,
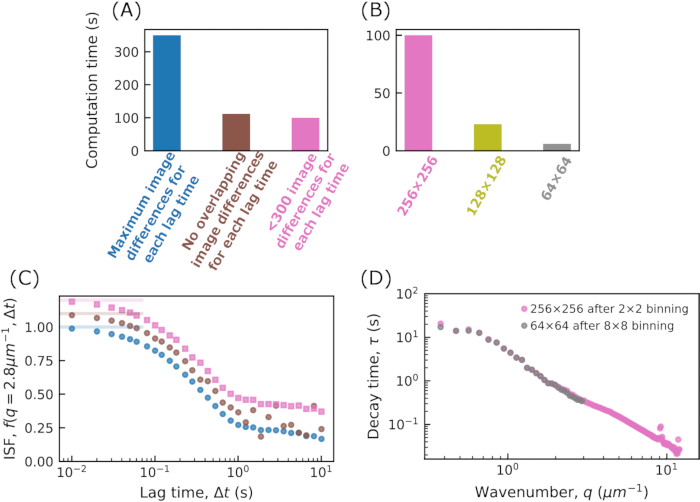
Figure 2: Computation time for calculating the DDM matrix. In (A) and (B) the time for calculating the DDM matrix,  , is shown. The data used in all cases is a movie of 5000 frames with an image size of 512 x 512 pixels. The DDM matrix was calculated for 30 lag times, logarithmically spaced between 1 frame (0.01 s) and 1000 frames (10 s). The code was run on an Intel i7-10700 2.90 GHz desktop computer with 32 GB RAM. In (A), the effect of varying how many image differences are used in computing the DDM matrix for each lag time is shown. For this, the images are binned to result in an image size of 256 x 256. For each lag time Δt, images separated by that Δt are subtracted and the resulting matrix is Fourier transformed. For a given Δt, all pairs of images separated by that Δt can be used (shown in blue), only non-overlapping image pairs can be used (e.g., frames 1 and 10, 10 and 19, etc.; shown in brown), or 300 image pairs or fewer can be used for each Δt. In (B), the effect of changing the image size on the computation time is shown. The images were binned either by grouping 2 x 2, 4 x 4, or 8 x 8 pixels, resulting in image sizes of 256 x 256, 128 x 128, or 64 x 64, respectively. For each, about 300 image pairs are used in computing the DDM matrix for each Δt. (C) From the DDM matrix, the intermediate scattering function (ISF) can be extracted. This is shown for the three cases in (A). The blue data points (with no offset) correspond to the ISF when the maximum number of image pairs are used for each Δt; the brown data points (with an offset of 0.1) correspond to the ISF when non-overlapping image pairs are used for each Δt; and the pink data points (with an offset of 0.2) correspond to the ISF when at most 300 image pairs are used for each Δt. The ISF found using non-overlapping image pairs shows noisiness at long Δt. For that case, few image pairs are used at long Δt (e.g., for Δt of 1000 frames, only 4 image pairs are used). (D) By fitting the ISF to an exponential function, the characteristic decay time, τ, for each wavenumber, q, is determined. In pink, results are shown after binning the original images by 2 x 2, resulting in an image size of 256 x 256. In gray, results are shown after binning by 8 x 8, resulting in an image size of 64 x 64. By binning the data, information about the dynamics at higher wavenumbers is lost, but calculating the DDM matrix for the 64 x 64 images is about 16x faster than for the 256 x 256 images. Please click here to view a larger version of this figure.
, is shown. The data used in all cases is a movie of 5000 frames with an image size of 512 x 512 pixels. The DDM matrix was calculated for 30 lag times, logarithmically spaced between 1 frame (0.01 s) and 1000 frames (10 s). The code was run on an Intel i7-10700 2.90 GHz desktop computer with 32 GB RAM. In (A), the effect of varying how many image differences are used in computing the DDM matrix for each lag time is shown. For this, the images are binned to result in an image size of 256 x 256. For each lag time Δt, images separated by that Δt are subtracted and the resulting matrix is Fourier transformed. For a given Δt, all pairs of images separated by that Δt can be used (shown in blue), only non-overlapping image pairs can be used (e.g., frames 1 and 10, 10 and 19, etc.; shown in brown), or 300 image pairs or fewer can be used for each Δt. In (B), the effect of changing the image size on the computation time is shown. The images were binned either by grouping 2 x 2, 4 x 4, or 8 x 8 pixels, resulting in image sizes of 256 x 256, 128 x 128, or 64 x 64, respectively. For each, about 300 image pairs are used in computing the DDM matrix for each Δt. (C) From the DDM matrix, the intermediate scattering function (ISF) can be extracted. This is shown for the three cases in (A). The blue data points (with no offset) correspond to the ISF when the maximum number of image pairs are used for each Δt; the brown data points (with an offset of 0.1) correspond to the ISF when non-overlapping image pairs are used for each Δt; and the pink data points (with an offset of 0.2) correspond to the ISF when at most 300 image pairs are used for each Δt. The ISF found using non-overlapping image pairs shows noisiness at long Δt. For that case, few image pairs are used at long Δt (e.g., for Δt of 1000 frames, only 4 image pairs are used). (D) By fitting the ISF to an exponential function, the characteristic decay time, τ, for each wavenumber, q, is determined. In pink, results are shown after binning the original images by 2 x 2, resulting in an image size of 256 x 256. In gray, results are shown after binning by 8 x 8, resulting in an image size of 64 x 64. By binning the data, information about the dynamics at higher wavenumbers is lost, but calculating the DDM matrix for the 64 x 64 images is about 16x faster than for the 256 x 256 images. Please click here to view a larger version of this figure.
6. Fitting the DDM matrix or the ISF
- Initialize an instance of the DDM_Fit class. To do so, pass to DDM_Fit the filename of the YAML file containing the image metadata and parameters for fitting.
- Decide which model for the DDM matrix or the ISF to use for fitting the data. List the available models by executing the function print_fitting_models. Specify the model to be used in the YAML parameter file or by using the function reload_fit_model_by_name.
- Set the initial guesses and bounds for each parameter in the chosen model in the provided YAML parameter file. To change the initial guess for any parameter, use the function set_parameter_initial_guess. Set bounds for the parameters with the function set_parameter_bounds. For example, as seen in Supplementary File 2, for the data of tracer beads in the vimentin network, the initial guess for the decay time was 1 s and the bounds on that parameter were 0.01 s and 2000 s.
- Execute the fit with the function fit. Assign a variable to the output of this function to easily access the results.
NOTE: This function can take many optional arguments. See the code documentation and provided examples for a list of such arguments and when to consider setting them to non-default values.
7. Interpreting the fit results
- Generate plots for inspecting the fits and the q-dependence of the fit parameters with the function fit_report.
NOTE: This function will generate a series of plots, which will also be saved as a PDF. Optional arguments to this function can be used to modify the plots produced. - Among the generated plots will be a figure with 2 x 2 subplots showing the DDM matrix or ISF (depending on the chosen fitting model) at four q-values (specified as an optional argument to fit_report), along with the calculated DDM matrix or ISF using the model and best fit parameters. To plot the DDM matrix or ISF along with the best fit in an interactive way, use the class Browse_DDM_Fits as shown in the provided examples when the Jupyter Notebook environment is used.
- From the plot of the characteristic decay time τ vs. the wavenumber q, determine whether the dynamics follow diffusive, subdiffusive, ballistic, or some other type of motion. This can be done by looking for the power law relationship between τ and q.
NOTE: On the log-log plot of τ vs. q generated by the function fit_report, three lines will be shown, corresponding to power law fits over a specified range of q values. The solid black line corresponds to fitting τ vs. q to a power law, τ = 1 / Kqβ, where K and β are free parameters. The dashed line in orange corresponds to fitting to simple diffusion, τ = 1 / Dq2, where D is a diffusion coefficient. The dot-dashed line in blue corresponds to fitting to τ = 1 / vq, where v is a velocity.
8. Saving the results
- The results of the fit will be saved in a xarray dataset. Use the xarray function to_netcdf or Python's built-in pickle module to save this data structure to disk. Use the xarray function open_dataset to load these netCDF files.
- Use the function save_fit_results_to_excel to save the fit results, along with the data, to a worksheet file.
Representative Results
Here, we show examples of the analysis done with PyDDM from two different sets of experiments. In one set of experiments, sub-micron tracer beads were embedded in networks consisting of the intermediate filament protein vimentin and imaged using a 100x objective lens in brightfield mode at 100 frames/s (Figure 3A). Vimentin is expressed in mesenchymal cells and is a key determinant of the mechanical properties of the cytoplasm65 and the mechanical stability of the nucleus in cells performing confined migration66,67. So far, reconstituted vimentin networks have been studied primarily by macroscopic rheology64,68,69, whereas the dynamics have received comparatively little attention13,70,71. Additional details of these experiments can be found in Supplementary File 2. In the other set of experiments, active cytoskeleton networks were prepared with actin, microtubules, and myosin. Spectrally distinct fluorescent labels allowed the actin and microtubule filaments to be imaged with a two-color laser-scanning confocal microscope using a 60x objective lens at 2.78 frames/s (Figure 3B,C). Actin and microtubule filaments are both important drivers of dynamic cell shape changes, with their actions coordinated by mechanical and biochemical interactions72. Additional details of these experiments can be found in11. Individual frames from image sequences taken in these experiments are shown in Figure 3.
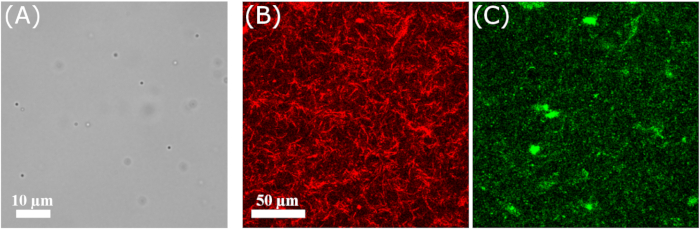
Figure 3: Images from the time series analyzed. (A) Brightfield image of 0.6 µm beads in a vimentin network. (B,C) Image of the (B) microtubules and (C) actin in an active actin-microtubule composite taken with a 60x objective on a laser-scanning confocal microscope, using 561 nm excitation light for the microtubule imaging and 488 nm excitation light for the actin imaging. Please click here to view a larger version of this figure.
For images of tracer beads in vimentin networks, movies of 5000 frames with a size of 512 x 512 pixels at 100 frames/s were recorded. From these, the DDM matrix was computed at 60 logarithmically spaced lag times between 1 and 1000 frames, or 0.01 s and 10 s. To estimate the background, B, the mean of the squared Fourier-transformed images, 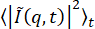 , was computed and set equal to
, was computed and set equal to 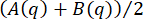 55,73. An assumption was made that, over the largest 10% of q-values, this quantity equals B/2 and that B is independent of q. This is the package's default method for estimating B, but other methods are possible by setting the background_method parameter to a different value.
55,73. An assumption was made that, over the largest 10% of q-values, this quantity equals B/2 and that B is independent of q. This is the package's default method for estimating B, but other methods are possible by setting the background_method parameter to a different value.
With the parameters A(q) and B determined from 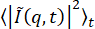 , one can extract the intermediate scattering function (ISF) from the DDM matrix. Example ISFs are shown in Figure 4. In Figure 4A, the ISF from images of 0.6 µm diameter beads embedded in a network with a vimentin concentration of 19 µM is shown. In Figure 4B, the ISF for the same type of beads in a network with a vimentin concentration of 34 µM is shown. Interestingly, in neither case did the ISF decay to zero. At large lag times, the ISF should approach zero for ergodic systems. That is, in such systems, density fluctuations should completely decorrelate over large lag times. The fact that the ISF here did not decay to zero could have resulted from inaccurate estimates of A(q) and B, which were used to find the ISF from the computed DDM matrix. Notably, the method employed here can overestimate B in certain scenarios62. However, it is more likely that the dynamics of the tracer beads are truly nonergodic as the beads have a comparable size to the network mesh size and may, therefore, become caged. Other data corroborated the finding of nonergodicity. Namely, the bead size, 0.6 µm, was larger than the calculated average value for the mesh sizes of 0.4 µm for the 19 µM concentration and 0.3 µm for the 34 µM concentration. Additionally, the results from single particle tracking of these tracer beads, which are shown later, also showed confined motion.
, one can extract the intermediate scattering function (ISF) from the DDM matrix. Example ISFs are shown in Figure 4. In Figure 4A, the ISF from images of 0.6 µm diameter beads embedded in a network with a vimentin concentration of 19 µM is shown. In Figure 4B, the ISF for the same type of beads in a network with a vimentin concentration of 34 µM is shown. Interestingly, in neither case did the ISF decay to zero. At large lag times, the ISF should approach zero for ergodic systems. That is, in such systems, density fluctuations should completely decorrelate over large lag times. The fact that the ISF here did not decay to zero could have resulted from inaccurate estimates of A(q) and B, which were used to find the ISF from the computed DDM matrix. Notably, the method employed here can overestimate B in certain scenarios62. However, it is more likely that the dynamics of the tracer beads are truly nonergodic as the beads have a comparable size to the network mesh size and may, therefore, become caged. Other data corroborated the finding of nonergodicity. Namely, the bead size, 0.6 µm, was larger than the calculated average value for the mesh sizes of 0.4 µm for the 19 µM concentration and 0.3 µm for the 34 µM concentration. Additionally, the results from single particle tracking of these tracer beads, which are shown later, also showed confined motion.
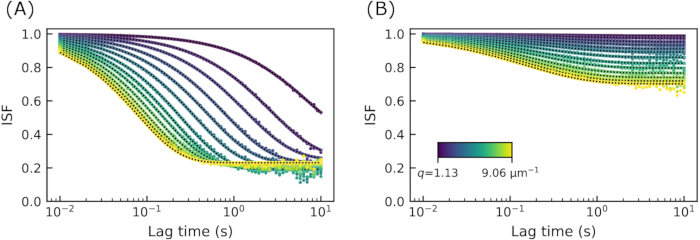
Figure 4: Intermediate scattering functions at several wavenumbers for vimentin networks. The ISF is plotted as a function of lag time for q values from about 1 to 9 µm-1. (A) The ISF from images of 0.6 µm beads in a vimentin network with vimentin concentration of 19 µM. (B) The ISF from images of 0.6 µm beads in a vimentin network with vimentin concentration of 34 µM. The long lag time plateau of the ISF at a value well above zero indicates nonergodicity. Please click here to view a larger version of this figure.
Given that the dynamics are likely nonergodic, the ISFs are fit to the form 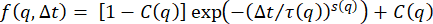 , where C is the nonergodicity factor 32. This form of the ISF has been used in previous studies of non-ergodic dynamics, such as that of colloidal gels32,74 or tracer particles in actin-microtubule networks 10. The dotted black lines in Figure 4 show the fits along with the data. From these fits, one can now look at the q-dependence of the decay time, τ, and of the nonergodicity parameter, C.
, where C is the nonergodicity factor 32. This form of the ISF has been used in previous studies of non-ergodic dynamics, such as that of colloidal gels32,74 or tracer particles in actin-microtubule networks 10. The dotted black lines in Figure 4 show the fits along with the data. From these fits, one can now look at the q-dependence of the decay time, τ, and of the nonergodicity parameter, C.
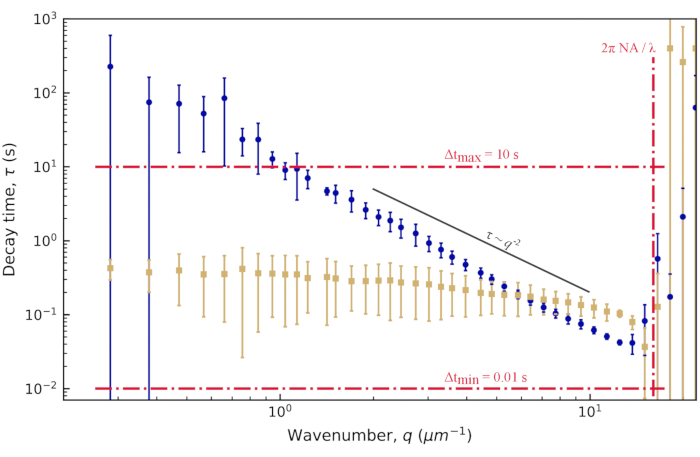
Figure 5: Decay time vs. wavenumber for vimentin networks. From fits to the ISF, the decay time τ is determined for a range of q values. For clarity, we are not showing the value of τ for every q, but just a logarithmically spaced set. In blue (tan) is the data from images of 0.6 µm beads within vimentin networks with a vimentin concentration of 19 µM (34 µM). The error bars represent the standard deviations in τ across multiple movies (four movies for the data with the 19 µM network [blue] and five movies for the data with the 34 µM network [tan]). Red dash-dotted lines mark estimated bounds for our temporal and spatial resolution, as described in the results. The solid black line shows 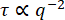 scaling, which would indicate diffusive motion. Neither data set follows this scaling. Rather, beads in the 19 µM network show subdiffusive motion (
scaling, which would indicate diffusive motion. Neither data set follows this scaling. Rather, beads in the 19 µM network show subdiffusive motion (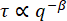 with β > 2), and beads in the 34 µM network show confined or caged motion. Please click here to view a larger version of this figure.
with β > 2), and beads in the 34 µM network show confined or caged motion. Please click here to view a larger version of this figure.
The decay times showed a large amount of uncertainty, both at the low q and high q extremes, as seen in Figure 5. The error bars on this plot show the standard deviation among four videos analyzed for the lower vimentin concentration case or five videos analyzed for the higher concentration. To understand the source of the large uncertainty at these extremes, consider both the temporal and spatial resolution. Approximate limits of the resolution are shown with three red dash-dotted lines. The two horizontal lines correspond to the minimum and maximum lag times probed. Given the frame rate of 100 frames/s and the maximum lag time corresponding to 1000 frames (20% of the total video duration), accuracy was lost when measuring dynamics occurring faster than 0.01 s or slower than 10 s. At the lower q-values, the fitted values for τ were greater than 10 s. Therefore, large uncertainties should be expected in decay times that are larger than the maximum lag time. At the higher end of the q-range, the decay time approached the minimum lag time of 0.01 s but remained above it. Rather than being limited by the temporal resolution, at these higher q values, the spatial resolution may be the limiting factor. Given the pixel size of 0.13 µm, the largest value for q was about 24 µm-1. However, the diffraction-limited resolution does not necessarily allow accurate measurements of the dynamics at these high spatial frequencies. Approximating the optical resolution as 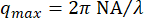 leads to an upper wavenumber limit of about 16 µm-1, given the objective lens's numerical aperture, NA, of 1.4 and wavelength of light,
leads to an upper wavenumber limit of about 16 µm-1, given the objective lens's numerical aperture, NA, of 1.4 and wavelength of light, 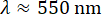 . This is demarked by the vertical red dash-dotted line in Figure 5. Indeed, the data were noisy at large values of q. Even before this approximate upper limit of q, increased uncertainty in τ was seen, and this could be from overestimating qmax. Poorer optical resolution than predicted may be because an oil immersion lens was used to image beyond the coverslip into an aqueous sample or because the condenser lens was imperfectly aligned.
. This is demarked by the vertical red dash-dotted line in Figure 5. Indeed, the data were noisy at large values of q. Even before this approximate upper limit of q, increased uncertainty in τ was seen, and this could be from overestimating qmax. Poorer optical resolution than predicted may be because an oil immersion lens was used to image beyond the coverslip into an aqueous sample or because the condenser lens was imperfectly aligned.
For the 0.6 µm beads embedded in the less concentrated network (19 µM vimentin), it can be observed from the log-log plot of the decay time vs. wavenumber that the decay time decreased with wavenumber in a way consistent with a power law (Figure 5). However, it does not seem to follow what would be expected for normal diffusive motion, where 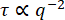 . Rather, τ decreased more steeply with increasing q. This is indicative of subdiffusive motion, which often occurs for beads in crowded environments such as these. Fitting τ(q) over the range of 1.4 µm-1 to 12.3 µm-1 to a power law of the form τ = 1/Kqβ yields the transport parameters K = 0.0953 μmβ / s and β = 2.2. For those more accustomed to thinking about normal diffusion vs. subdiffusion in terms of the mean squared displacement (MSD) of tracer particles as a function of lag time (i.e., MSD = K' Δtα), it is helpful to recognize that the subdiffusive scaling exponent in the MSD equation, α, is equivalent to α = 2 / β. In other words, the value of β = 2.2 is consistent with a subdiffusive scaling exponent in the MSD equation of α = 0.9. One would set PyDDM to fit τ(q) over this range of q-values by specifying the indices of the array of q with either the parameter Good_q_range in the YAML file or by passing the optional argument forced_qs to the function generate_fit_report. The range of q from 1.4 µm-1 to 12.3 µm-1 would, for the data here, correspond to indices of the array of q from 15 to 130.
. Rather, τ decreased more steeply with increasing q. This is indicative of subdiffusive motion, which often occurs for beads in crowded environments such as these. Fitting τ(q) over the range of 1.4 µm-1 to 12.3 µm-1 to a power law of the form τ = 1/Kqβ yields the transport parameters K = 0.0953 μmβ / s and β = 2.2. For those more accustomed to thinking about normal diffusion vs. subdiffusion in terms of the mean squared displacement (MSD) of tracer particles as a function of lag time (i.e., MSD = K' Δtα), it is helpful to recognize that the subdiffusive scaling exponent in the MSD equation, α, is equivalent to α = 2 / β. In other words, the value of β = 2.2 is consistent with a subdiffusive scaling exponent in the MSD equation of α = 0.9. One would set PyDDM to fit τ(q) over this range of q-values by specifying the indices of the array of q with either the parameter Good_q_range in the YAML file or by passing the optional argument forced_qs to the function generate_fit_report. The range of q from 1.4 µm-1 to 12.3 µm-1 would, for the data here, correspond to indices of the array of q from 15 to 130.
For the 0.6 µm beads in the more concentrated network (34 µM), the decay time showed little dependence on q. This is likely due to the nonergodicity of beads in a network with a smaller mesh size. To probe the nonergodicity in this system, the nonergodicity parameter, C, should be plotted as a function of q, as in Figure 6. For the 0.6 µm beads in the 19 µM vimentin network, C ≈ 0.2 with little dependence on q (not shown). However, for the network with 34 µM vimentin and for a network with an even higher concentration of 49 µM vimentin, the log of C was proportional to q2 as shown in Figure 6. This relationship between C and q is expected for confined motion. For beads trapped within pockets of the network, the MSD is expected to plateau at long enough lag times (i.e., 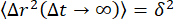 , where
, where 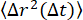 is the MSD and δ2 is the maximum MSD). Since the ISF can be expressed in terms of the MSD as
is the MSD and δ2 is the maximum MSD). Since the ISF can be expressed in terms of the MSD as 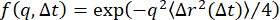 , and since the nonergodic ISF goes to C at long lag times (i.e.,
, and since the nonergodic ISF goes to C at long lag times (i.e., 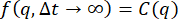 ), the relationship
), the relationship 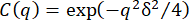 is obtained32,75. Therefore, one can use C(q) to find δ2, and this yielded δ2 = 0.017 μm2 and 0.0032 μm2 for the 34 and 49 µM vimentin networks, respectively (corresponding to δ = 0.13 μm and 0.057 μm).
is obtained32,75. Therefore, one can use C(q) to find δ2, and this yielded δ2 = 0.017 μm2 and 0.0032 μm2 for the 34 and 49 µM vimentin networks, respectively (corresponding to δ = 0.13 μm and 0.057 μm).
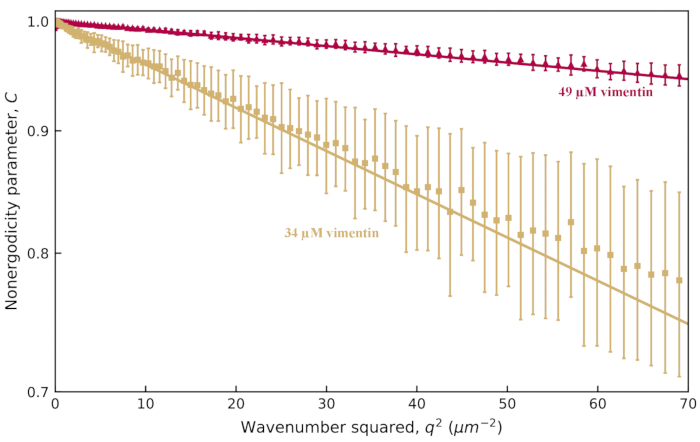
Figure 6: Nonergodicity parameter vs. wavenumber for vimentin networks. From fits to the ISF, the nonergodicity parameter C is determined for a range of q values. In tan (red) is the data from images of 0.6 µm beads within vimentin networks with a vimentin concentration of 34 µM (49 µM). The error bars represent the standard deviations in τ across multiple movies (five movies for the data with the 34 µM network [tan] and four movies for the data with the 49 µM network [red]). The y-axis has logarithmic scaling. One observes a q-dependence of C that follows 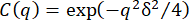 , which allows for extracting the maximum mean squared displacement, δ2. Fits to
, which allows for extracting the maximum mean squared displacement, δ2. Fits to 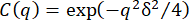 are shown with the solid lines. Please click here to view a larger version of this figure.
are shown with the solid lines. Please click here to view a larger version of this figure.
One can use other methods to extract the confinement size δ from the data as well as the subdiffusive exponent found from examining τ(q) for beads within the 19 µM vimentin network. Firstly, one can use the method described by Bayles et al.76 and Edera et al.77 to extract the MSD from the DDM matrix. Notably, this method requires no fitting of the DDM matrix. One only needs to compute the DDM matrix, D(q, Δt), and 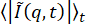 (from which A(q) and B can be determined). Then, to find the MSD, one uses the relationship
(from which A(q) and B can be determined). Then, to find the MSD, one uses the relationship 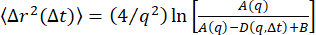 . Note that this method to find the MSD assumes that the distribution of particle displacements is Gaussian, though previous work has shown that, in certain cases, MSDs derived from DDM do agree with MSDs from particle tracking, even when the displacements are non-Gaussian73. For this system, as expected78, there is non-Gaussianity in the distribution of large displacements, as seen in Figure S1. In the PyDDM package, the function extract_MSD should be executed, which returns
. Note that this method to find the MSD assumes that the distribution of particle displacements is Gaussian, though previous work has shown that, in certain cases, MSDs derived from DDM do agree with MSDs from particle tracking, even when the displacements are non-Gaussian73. For this system, as expected78, there is non-Gaussianity in the distribution of large displacements, as seen in Figure S1. In the PyDDM package, the function extract_MSD should be executed, which returns  . Secondly, one can use single particle tracking to find the MSD. Though DDM can be used to analyze images where either the high density of particles or the limited optical resolution prohibits accurate particle localization, for the images of 0.6 µm beads in vimentin networks, we were able to localize and track beads using the trackpy software (https://github.com/soft-matter/trackpy)79. This particle tracking software package uses the algorithms described by Crocker and Grier80.
. Secondly, one can use single particle tracking to find the MSD. Though DDM can be used to analyze images where either the high density of particles or the limited optical resolution prohibits accurate particle localization, for the images of 0.6 µm beads in vimentin networks, we were able to localize and track beads using the trackpy software (https://github.com/soft-matter/trackpy)79. This particle tracking software package uses the algorithms described by Crocker and Grier80.
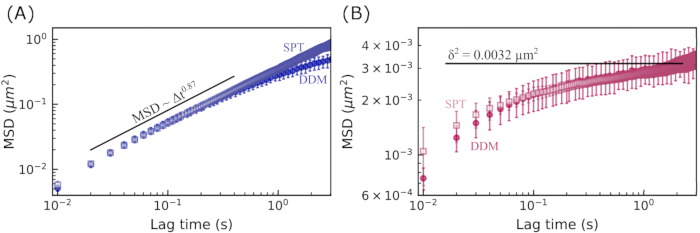
Figure 7: Mean squared displacement vs. lag time for vimentin networks. The MSD was determined using two methods. First, the MSD was computed from the DDM matrix (shown with solid symbols). Next, the MSD was determined by using single-particle tracking (SPT) to find particle trajectories (open symbols). Error bars are determined in the same way as described in the previous two figure legends. (A) MSDs for 0.6 µm beads in the 19 µM vimentin network indicate subdiffusive motion, with good agreement between the two methods of finding the MSD. (B) MSDs for 0.6 µm beads in the 49 µM vimentin network indicate caged motion, with good agreement between the two methods of finding the MSD and with the maximum MSD found from the nonergodicity parameter. Please click here to view a larger version of this figure.
The MSDs vs. lag time for 0.6 µm beads in the 19 µM vimentin network and in the 49 µM vimentin network are shown in Figure 7. In both cases, the MSD determined from DDM agreed well with the MSD found through single-particle tracking (SPT). Furthermore, for the less concentrated network, the subdiffusive scaling exponent (α di 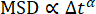 ) was about 0.9. This is consistent with the τ(q) scaling of
) was about 0.9. This is consistent with the τ(q) scaling of 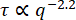 found by fitting the ISF to determine τ(q) (that is, 2/2.2 = 0.9). For the more concentrated network, the MSD plateaus at longer lag times. The maximum MSD found by analyzing the q-dependence of the nonergodicity parameter (shown in Figure 7B with the horizontal line at δ2 = 0.0032 μm2) was approximately the same value that the MSDs from both SPT and DDM seemed to be plateauing toward. There is a discrepancy between the longest lag time MSDs determined from DDM and SPT in Figure 7A. While this may be due to a limited number of long lag time trajectories, it may also be the case that further optimizing the range of q values for which the DDM matrix is used to estimate
found by fitting the ISF to determine τ(q) (that is, 2/2.2 = 0.9). For the more concentrated network, the MSD plateaus at longer lag times. The maximum MSD found by analyzing the q-dependence of the nonergodicity parameter (shown in Figure 7B with the horizontal line at δ2 = 0.0032 μm2) was approximately the same value that the MSDs from both SPT and DDM seemed to be plateauing toward. There is a discrepancy between the longest lag time MSDs determined from DDM and SPT in Figure 7A. While this may be due to a limited number of long lag time trajectories, it may also be the case that further optimizing the range of q values for which the DDM matrix is used to estimate 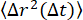 for each lag time (as done by Bayles et al.76 and Edera et al.77) would improve our results, and such optimization will be the focus of future work.
for each lag time (as done by Bayles et al.76 and Edera et al.77) would improve our results, and such optimization will be the focus of future work.
These experiments where image sequences were recorded of tracer beads embedded in a network of vimentin intermediate filaments allowed for independent analyses: DDM (using the package described here) and SPT (using trackpy). Both analyses can reveal the degree of subdiffusion and confinement length, allowing one to use two independent image analysis techniques to provide complementary metrics. There are additional quantities one can compare from SPT and DDM. For example, heterogeneity in the dynamics of the sample can reveal itself as non-Gaussianity in the distribution of particle displacements (i.e., the van Hove distribution) determined from SPT, as well as in an ISF determined from DDM that fits to a stretched exponential34,35. Figure S1 shows the van Hove distribution for the 0.6 µm particles in vimentin networks and discusses the stretching exponent found from fitting the ISFs—metrics used in tandem in previous studies to demonstrate the heterogeneous dynamics of particles within biomimetic systems9,10,47 or other crowded environments 34. As another example, the ISF can be calculated from particle trajectories measured with SPT and compared with the DDM-acquired ISFs. While the mean squared displacements and displacement distributions are the metrics most often pulled from SPT analysis, one can also compute the ISF from particle trajectories, 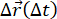 , using
, using 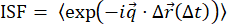 (see Figure S2). This ISF can be compared with DDM-generated ISFs and used to reveal dynamics not apparent in the MSD59.
(see Figure S2). This ISF can be compared with DDM-generated ISFs and used to reveal dynamics not apparent in the MSD59.
While acquiring images of tracer particles within a network may allow one to use the complementary analysis methods of SPT and DDM, it is important to note that an advantage of DDM over SPT is that it does not require images of beads (or other features) that can be easily localized and tracked. To demonstrate this point, we next highlight the analysis of active networks of actin and microtubule filaments, where fluorescent labeling of actin and tubulin allows for the imaging of both filament types, distinguished from each other via different fluorophores, with a multi-color laser-scanning confocal microscope.
Images were acquired with a laser-scanning confocal microscope of actin-microtubule networks with activity driven by myosin (rabbit skeletal muscle myosin II; Cytoskeleton #MY02). Details of the experiments and results have been previously described11, and the representative results shown here are from the analysis of two movies provided in the supplemental materials (movies S1 and S4) for11. Both image sequences were recorded at 2.78 frames/s for 1000 frames.
To analyze these images, the DDM matrix was calculated for 50 lag times ranging from 0.4 s to 252 s (1 frame to 700 frames). The DDM matrix was then fit to the model  , with the intermediate scattering function being
, with the intermediate scattering function being 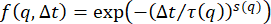 . There are, therefore, four fitting parameters: A, τ, s, and B. The results of these fits are shown in Figure 8. It was observed that the DDM matrix for a particular q-value had a plateau at low lag times, increased with lag time, and then plateaued (or showed signs of beginning to plateau) at large lag times. The DDM matrix for the lower values of q did not reach a plateau at long lag times. One should, therefore, expect poor accuracy in the measurement of the decay time for these low q (large length scale) dynamics.
. There are, therefore, four fitting parameters: A, τ, s, and B. The results of these fits are shown in Figure 8. It was observed that the DDM matrix for a particular q-value had a plateau at low lag times, increased with lag time, and then plateaued (or showed signs of beginning to plateau) at large lag times. The DDM matrix for the lower values of q did not reach a plateau at long lag times. One should, therefore, expect poor accuracy in the measurement of the decay time for these low q (large length scale) dynamics.
The characteristic decay times, τ, from the fits to the DDM matrix are shown in Figure 9. Results are presented for an active actin-microtubule composite network (similar to movie S111) and for an active actin network (similar to movie S411). Both networks were prepared with the same concentrations of actin and myosin, but the actin-only network was created without tubulin, as described in11. For these two types of active networks, the observed power law relationship was 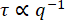 . This scaling indicates ballistic motion and that the myosin-driven contraction and flow dominate over the thermal motion of the filaments. From τ = (vq)-1, a characteristic velocity, v, of about 10 nm/s for the active actin-microtubule network and 75 nm/s for the active actin network could be found. These values are consistent with the particle image velocimetry analysis of the same videos shown in11. The
. This scaling indicates ballistic motion and that the myosin-driven contraction and flow dominate over the thermal motion of the filaments. From τ = (vq)-1, a characteristic velocity, v, of about 10 nm/s for the active actin-microtubule network and 75 nm/s for the active actin network could be found. These values are consistent with the particle image velocimetry analysis of the same videos shown in11. The 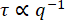 scaling did not hold at the lower q values for the active actin-microtubule composite network. This is likely because the true decay times for this actin-microtubule composite network at the lower q values are longer than the maximum lag time of the computed DDM matrix. The maximum lag time is indicated with the horizontal red line in Figure 9, and the decay times deviated from the expected
scaling did not hold at the lower q values for the active actin-microtubule composite network. This is likely because the true decay times for this actin-microtubule composite network at the lower q values are longer than the maximum lag time of the computed DDM matrix. The maximum lag time is indicated with the horizontal red line in Figure 9, and the decay times deviated from the expected 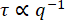 scaling near these longer times.
scaling near these longer times.
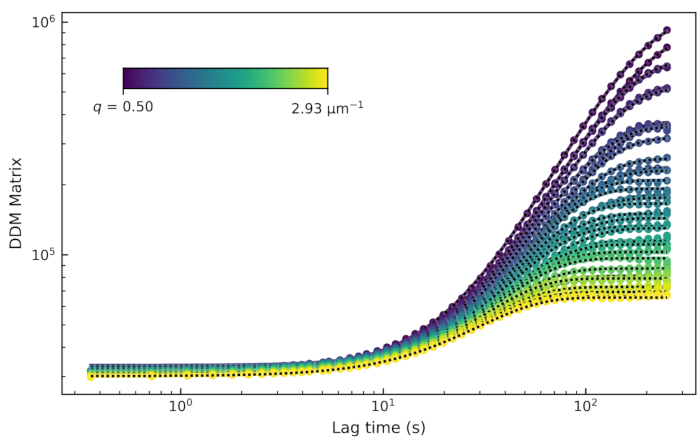
Figure 8: DDM matrix vs. lag time for an active actin-microtubule composite network. The DDM matrix for several values of q is plotted as a function of lag time from a movie of a composite network composed of 2.9 µM actin monomers, 2.9 µM tubulin dimers, and 0.24 µM myosin. These data show the analysis of just the microtubule channel of a multicolor time series of images. Please click here to view a larger version of this figure.
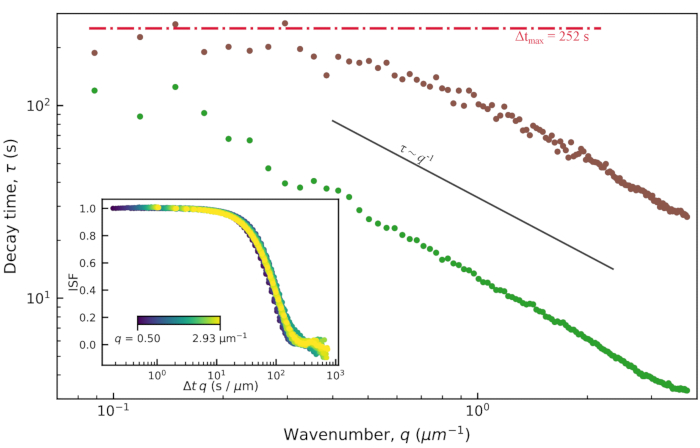
Figure 9: Decay time vs. wavenumber for active actin-microtubule networks. From fitting the DDM matrix, the decay time, τ, as a function of wavenumber, q, is found. Plotted is τ vs q for images of an active actin-microtubule network (analyzing just the microtubule channel) in brown and for images of an active actin network in green. Both networks have the same concentrations of actin and myosin (2.9 µM and 0.24 µM, respectively); the actin-microtubule composite has 2.9 µM of tubulin dimers. The decay times for the active actin network are much smaller than the decay times for the active actin-microtubule network, which indicates faster motion of the active actin network. In both cases, the dynamics are ballistic as the data follows a 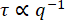 trend. Inset: the plot of the ISFs vs. the lag time scaled by the wavenumber (Δt × q) shows a collapse of the ISFs over a range of q values. This also indicates ballistic motion. The ISFs shown in this inset are from the active actin network. Please click here to view a larger version of this figure.
trend. Inset: the plot of the ISFs vs. the lag time scaled by the wavenumber (Δt × q) shows a collapse of the ISFs over a range of q values. This also indicates ballistic motion. The ISFs shown in this inset are from the active actin network. Please click here to view a larger version of this figure.
For this data of active networks, we chose to fit the DDM matrix,  . This contrasts with what was done for the data of beads in the vimentin network, where A (q) and B were estimated without any fitting to isolate the ISF, f (q, Δt). In this case, for the active network data, A and B were left as fitting parameters because the methods used to estimate B did not result in good fits. The default method to estimate B is to compute
. This contrasts with what was done for the data of beads in the vimentin network, where A (q) and B were estimated without any fitting to isolate the ISF, f (q, Δt). In this case, for the active network data, A and B were left as fitting parameters because the methods used to estimate B did not result in good fits. The default method to estimate B is to compute 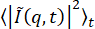 and to assume that, at large q, this goes to B/2. However, this method overestimated B for this data, which was seen in the fact that, when calculating the ISFs from B estimated in this way (not shown), the ISFs were greater than 1 at early lag times (whereas they should go from a maximum of 1 to either zero or some nonergodicity parameter with increasing lag time). One can select other methods for estimating B using the parameter background_method. One of these other methods is to estimate B to be the minimum of the DDM matrix at early lag times (set with background_method=1). A similar method was used by Bayles et al.76, though they did not assume B was constant with q. Another option is to estimate B to be the average value over all lag times of the DDM matrix at the maximum q (set with background_method=2). These different methods for estimating the background, as well as the results for allowing B to be a free fitting parameter, are shown in Figure 10. From those plots, one can see that the amplitude, A, did not reach zero at the largest q values probed, since
and to assume that, at large q, this goes to B/2. However, this method overestimated B for this data, which was seen in the fact that, when calculating the ISFs from B estimated in this way (not shown), the ISFs were greater than 1 at early lag times (whereas they should go from a maximum of 1 to either zero or some nonergodicity parameter with increasing lag time). One can select other methods for estimating B using the parameter background_method. One of these other methods is to estimate B to be the minimum of the DDM matrix at early lag times (set with background_method=1). A similar method was used by Bayles et al.76, though they did not assume B was constant with q. Another option is to estimate B to be the average value over all lag times of the DDM matrix at the maximum q (set with background_method=2). These different methods for estimating the background, as well as the results for allowing B to be a free fitting parameter, are shown in Figure 10. From those plots, one can see that the amplitude, A, did not reach zero at the largest q values probed, since 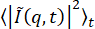 did not plateau at large q (Figure 10B), and since D(qmax, Δt) went from a lower lag time plateau to some higher lag time plateau (i.e., at qmax, there was a non-zero A; Figure 10D). Therefore, neither estimating B as
did not plateau at large q (Figure 10B), and since D(qmax, Δt) went from a lower lag time plateau to some higher lag time plateau (i.e., at qmax, there was a non-zero A; Figure 10D). Therefore, neither estimating B as 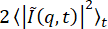 nor as
nor as 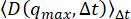 would be appropriate. One should inspect
would be appropriate. One should inspect 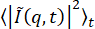 vs. q and D(qmax, Δt) vs. Δt before deciding on how (or if) to estimate B.
vs. q and D(qmax, Δt) vs. Δt before deciding on how (or if) to estimate B.
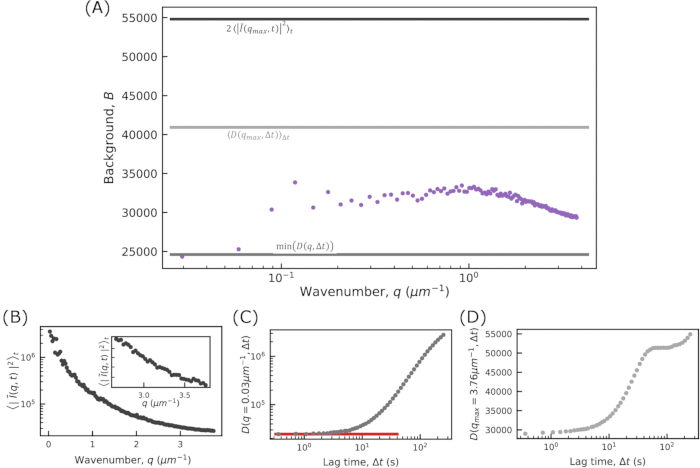
Figure 10: Background vs. wavenumber for active actin-microtubule networks. From fitting the DDM matrix, one can find the background, B, as a function of wavenumber, q. Shown is B vs. q for images of an active actin-microtubule network (analyzing just the microtubule channel) determined from these fits with the purple symbols. The three solid lines in (A) show estimates of the background found without any fitting. The top, darkest line in (A) shows the estimated background using 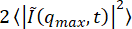 , which may be appropriate if
, which may be appropriate if 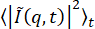 plateaus to a constant value at large q. From (B), note that
plateaus to a constant value at large q. From (B), note that 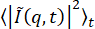 has yet to reach a constant value at the largest q probed. Therefore, using this method overestimates the background. The bottom line in (A) shows the estimated background using
has yet to reach a constant value at the largest q probed. Therefore, using this method overestimates the background. The bottom line in (A) shows the estimated background using 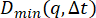 . If the DDM matrix shows a low lag time plateau as shown in (C) with the red line, then this method may be appropriate for estimating the background. The middle, lightest line in (A) shows the estimated background from
. If the DDM matrix shows a low lag time plateau as shown in (C) with the red line, then this method may be appropriate for estimating the background. The middle, lightest line in (A) shows the estimated background from 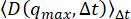 . This method may be appropriate if, at qmax, the amplitude, A, has reached zero. From (D), it is seen that the amplitude is non-zero and, therefore, this method overestimates the background. Please click here to view a larger version of this figure.
. This method may be appropriate if, at qmax, the amplitude, A, has reached zero. From (D), it is seen that the amplitude is non-zero and, therefore, this method overestimates the background. Please click here to view a larger version of this figure.
Supplementary Figure S1: Probability distributions of particle displacements. Probability distributions of particle displacements show non-Gaussianity for vimentin concentrations of 34 µM and 49 µM. Single-particle tracking of 0.6 µm diameter beads was performed in vimentin networks of different concentrations. Different lag times are shown in the displacement distributions for the three conditions. (A) The distribution of particle displacements in a 19 µM vimentin network are fit with a Gaussian function. The width of the Gaussian increases with increasing lag time. (B) The distribution of particle displacements in a 34 µM vimentin network shows more non-Gaussianity, especially at large displacements, than for the 19 µM case. (C) The distribution of particle displacements in a 49 µM vimentin network also shows non-Gaussianity. Further, the widths of the distributions do not increase with lag time as significantly as in the samples with lower vimentin concentrations, indicating confined motion. Non-Gaussian van Hove distributions (seen for all vimentin samples but most apparent in the higher concentrations) are associated with heterogeneous dynamics as often seen in the transport of particles in crowded and confined environments. Another indicator of heterogeneous transport that is determined from DDM analysis is the stretching exponent used to fit the intermediate scattering function (the parameter s in the equation for the ISF used here: 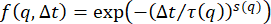 +
+ 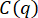 ). The average stretching exponents over the q-range of 0.4 µm-1 to 9.4 µm-1 are, from highest vimentin concentration to lowest, 0.53 ± 0.07, 0.64 ± 0.02, and 0.86 ± 0.04 (mean ± standard deviation). Please click here to download this File.
). The average stretching exponents over the q-range of 0.4 µm-1 to 9.4 µm-1 are, from highest vimentin concentration to lowest, 0.53 ± 0.07, 0.64 ± 0.02, and 0.86 ± 0.04 (mean ± standard deviation). Please click here to download this File.
Supplementary Figure S2: The intermediate scattering functions from DDM and SPT. The intermediate scattering functions (ISF) for five different wavenumbers are shown. The ISF versus lag time found through DDM is plotted with circular markers, and the ISF computed from single-particle trajectories with open squares. Dotted black lines show the fits to the DDM-acquired ISFs. The ISF is calculated from single-particle trajectories, 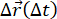 , using
, using 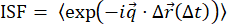 . In (A), the ISF is shown for 0.6 µm particles in the 19 µM vimentin networks. In (B), the ISF is shown for 0.6 µm particles in the 34 µM vimentin networks. The discrepancies in the ISF found from DDM and SPT are likely due to a limited number of long lag time trajectories. Please click here to download this File.
. In (A), the ISF is shown for 0.6 µm particles in the 19 µM vimentin networks. In (B), the ISF is shown for 0.6 µm particles in the 34 µM vimentin networks. The discrepancies in the ISF found from DDM and SPT are likely due to a limited number of long lag time trajectories. Please click here to download this File.
Supplementary File 1: Protocol for using DDM. The input and output of the steps shown in the protocol are presented. Please click here to download this File.
Supplementary File 2: Details of sample preparation and example parameter files for vimentin networks. Detailed steps for sample preparation and image acquisition on vimentin networks are provided. Additionally, an example parameters file for the analysis of data presented in the representative results section on vimentin networks is also provided. Please click here to download this File.
Discussion
The software package described here uses DDM to analyze density fluctuations observed in images acquired using an optical microscope. Representative results from the data of tracer particles embedded in vimentin networks were first shown. The analysis of such data can be used to characterize the mesh size and stiffness of the network similarly to how single-particle tracking has been used in many past studies of cytoskeleton networks6,12,13. An advantage of using DDM over single-particle tracking is that DDM does not require the particles to be localized. Therefore, even in images where the particle density is too high or the particles too small to localize and track, DDM can still determine the dynamics. Where single-particle tracking would be advantageous is when inspecting particle-to-particle variability. With DDM, one finds the ensemble averaged dynamics whereas, with single-particle tracking, one may compute both a single particle's MSD and the ensemble-averaged MSD. However, DDM can be used to investigate heterogeneous dynamics by analyzing multiple regions of interest within a large field of view.
Next, representative results from data of fluorescently labeled filaments in an active network composed of two differently labeled cytoskeletal filament types were shown11. With this data, the ballistic motion was characterized without needing any localizable features within the image. Since DDM extracts the ensemble averaged dynamics with few user inputs, it makes comparing image series acquired with different conditions straightforward (e.g., comparing samples with different ratios of actin to microtubules or samples with different concentrations of myosin, as done in50). Additionally, using fluorescent imaging, we can investigate the dynamics of different components of a network using multicolor labeling. This was done in11,50, where the dynamics of actin and of microtubules were separately analyzed in an active actin-microtubule composite network using multicolor imaging. In the representative results section here, just the results from the microtubule channel were shown, but in previous work, we compared the dynamics of the microtubule and actin filaments11.
We note that these representative results showcase either passive subdiffusion or active ballistic motion. Importantly, DDM can be used to analyze systems where there is a crossover in the type of dynamics at intermediate time or length scales. As examples, Kurzthaler et al. used DDM with a system of active Janus colloids to explore active directed motion at short time scales and randomization of the orientation at longer time scales59; Giavazzi et al. used DDM with a coarsening foam and found a crossover in the dynamics corresponding to the length scale of a bubble33; and Cho et al. used DDM with colloidal gels and found three distinguishable regimes at different length scales spanning from the fractal clusters to the whole network32.
The data included in the representative results section were acquired with brightfield microscopy and laser-scanning confocal microscopy. However, as previously noted, DDM can be used with many imaging modalities. With any imaging modality, users should consider optical settings such as the degree of optical sectioning or the depth of field. A high degree of optical sectioning may reduce signal from out-of-focus objects, but one will not be able to accurately measure dynamics over timescales greater than the timescale for objects to move out of the depth of field25,28. A more thorough discussion of how the q-dependent depth of field affects DDM analysis can be found in22. For brightfield imaging, users may also need to consider the sample thickness. While for weakly scattering samples, thicker samples may provide more signal42, turbid samples may require modifying the analysis to account for multiple scattering81. Finally, for imaging methods that are not linear space invariant (that is, where the intensity recorded by the camera of an object depends on where that object is in the x-y sample plane), one may need to account for the linear space variance, as demonstrated with dark-field DDM27.
For those getting started with DDM, we wish to emphasize the importance of considering the spatial and temporal resolution. When inspecting the determined decay times as a function of wavenumber, it is important to mark the limits of one's resolution (i.e., the maximum and minimum lag times and the maximum wavenumber, as done in Figure 5). One should think carefully about these limits prior to collecting data so that the optimal objective lens, image size, frame rate, and movie duration can be selected. The other important consideration is how to estimate the background parameter B. Multiple methods for estimating the background have been used in the literature, and the effects of over- or underestimating B have been described in prior publications62,77. As shown in Figure 10, PyDDM allows users to implement different methods for estimating B, and we suggest new users try these methods and evaluate which are appropriate to use.
A strength of this package is its thorough documentation and walkthroughs featuring example data, the storage and organization of metadata to keep track of how analyses were performed, and the flexibility in how to analyze the DDM matrix (various fitting models, multiple methods for estimating the background parameter B, the ability to find the MSD). However, there are multiple aspects to this code that could be improved. Currently, the code has not been optimized for fast computation speed. Methods for speeding up the computation have been reported61,62, and these will be implemented in future releases. Additionally, we plan to implement recently reported methods to better estimate uncertainties and to employ simulations for guiding users to the appropriate ISF model62. For other improvements, we hope users will contact us with suggestions.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
Parts of this research were funded by a National Institutes of Health R15 Award (National Institute of General Medical Sciences award no. R15GM123420, awarded to R.M.R.-A. and R.J.M.), a Cottrell Scholar Award from the Research Corporation for Science Advancement (award no. 27459, awarded to R.J.M.), and a William M. Keck Foundation Research Grant (awarded to R.M.R-A.). GHK gratefully acknowledges financial support from the Dutch Research Council (NWO; project number VI.C.182.004 of the NWO Talent Programme).
Materials
| CMOS camera, Orca-Flash 4.0 | Hamatsu | ||
| F-127 Pluronic | Sigma Aldrich | ||
| Jupyter Notebook | |||
| Nanodrop | Thermo Fisher | ||
| Nikon Ti-Eclipse microscope | Nikon | ||
| PLL-PEG-bio | SuSos AG, Dübendorf, Switzerland | ||
| Polystyrene beads | Sigma Aldrich | ||
| Protein dialysis mini-cassette | Thermo Fisher | ||
| PyDDM | University of San Diego | N/A | Open source software available from https://github.com/rmcgorty/PyDDM |
Riferimenti
- Burla, F., Mulla, Y., Vos, B. E., Aufderhorst-Roberts, A., Koenderink, G. H. From mechanical resilience to active material properties in biopolymer networks. Nature Reviews Physics. 1 (4), 249-263 (2019).
- Amblard, F., Maggs, A. C., Yurke, B., Pargellis, A. N., Leibler, S. Subdiffusion and anomalous local viscoelasticity in actin networks. Physical Review Letters. 77 (21), 4470-4473 (1996).
- Mizuno, D., Tardin, C., Schmidt, C. F., MacKintosh, F. C. Nonequilibrium mechanics of active cytoskeletal networks. Science. 315 (5810), 370-373 (2007).
- Bendix, P. M. et al. A quantitative analysis of contractility in active cytoskeletal protein networks. Biophysical Journal. 94 (8), 3126-3136 (2008).
- Pelletier, V., Gal, N., Fournier, P., Kilfoil, M. L. Microrheology of microtubule solutions and actin-microtubule composite networks. Physical Review Letters. 102 (18), 188303 (2009).
- Stuhrmann, B., Soares e Silva, M., Depken, M., MacKintosh, F. C., Koenderink, G. H. Nonequilibrium fluctuations of a remodeling in vitro cytoskeleton. Physical Review E. 86 (2), 020901 (2012).
- Sanchez, T., Chen, D. T. N., DeCamp, S. J., Heymann, M., Dogic, Z. Spontaneous motion in hierarchically assembled active matter. Nature. 491 (7424), 431-434 (2012).
- Alvarado, J., Sheinman, M., Sharma, A., MacKintosh, F. C., Koenderink, G. H. Molecular motors robustly drive active gels to a critically connected state. Nature Physics. 9 (9), 591-597 (2013).
- Anderson, S. J. et al. Filament rigidity vies with mesh size in determining anomalous diffusion in cytoskeleton. Biomacromolecules. 20 (12),4380-4388 (2019).
- Anderson, S. J., Garamella, J., Adalbert, S., McGorty, R. J., Robertson-Anderson, R. M. Subtle changes in crosslinking drive diverse anomalous transport characteristics in actin-microtubule networks. Soft Matter. 17 (16), 4375-4385 (2021).
- Lee, G. et al. Myosin-driven actin-microtubule networks exhibit self-organized contractile dynamics. Science Advances. 7 (6), eabe4334 (2021).
- Wong, I. Y. et al. Anomalous diffusion probes microstructure dynamics of entangled F-actin networks. Physical Review Letters. 92 (17), 178101 (2004).
- Köster, S., Lin, Y.-C., Herrmann, H., Weitz, D. A. Nanomechanics of vimentin intermediate filament networks. Soft Matter. 6 (9), 1910-1914 (2010).
- Chandrakar, P. et al. Engineering stability, longevity, and miscibility of microtubule-based active fluids. Soft Matter. 18 (9), 1852-1835 (2022).
- Alvarado, J., Cipelletti, L., H. Koenderink, G. Uncovering the dynamic precursors to motor-driven contraction of active gels. Soft Matter. 15 (42), 8552-8565 (2019).
- Linsmeier, I. et al. Disordered actomyosin networks are sufficient to produce cooperative and telescopic contractility. Nature Communications. 7 (1), 12615 (2016).
- Stam, S. et al. Filament rigidity and connectivity tune the deformation modes of active biopolymer networks. Proceedings of the National Academy of Sciences. 114 (47), E10037-E10045 (2017).
- Malik-Garbi, M. et al. Scaling behaviour in steady-state contracting actomyosin networks. Nature Physics. 15 (5), 509-516 (2019).
- Berezney, J., Goode, B. L., Fraden, S., Dogic, Z. Extensile to contractile transition in active microtubule-actin composites generates layered asters with programmable lifetimes. Proceedings of the National Academy of Sciences. 119 (5), e2115895119 (2022).
- Roostalu, J., Rickman, J., Thomas, C., Nédélec, F., Surrey, T. Determinants of polar versus nematic organization in networks of dynamic microtubules and mitotic motors. Cell. 175 (3), 796-808.e714 (2018).
- Cerbino, R., Trappe, V. Differential dynamic microscopy: probing wave vector dependent dynamics with a microscope. Physical Review Letters. 100 (18), 188102 (2008).
- Giavazzi, F., Brogioli, D., Trappe, V., Bellini, T., Cerbino, R. Scattering information obtained by optical microscopy: differential dynamic microscopy and beyond. Physical Review E. 80 (3), 031403 (2009).
- Giavazzi, F., Cerbino, R. Digital Fourier microscopy for soft matter dynamics. Journal of Optics. 16 (8), 083001 (2014).
- He, K., Spannuth, M., Conrad, J. C., Krishnamoorti, R. Diffusive dynamics of nanoparticles in aqueous dispersions. Soft Matter. 8 (47), 11933-11938 (2012).
- Lu, P. J. et al. Characterizing concentrated, multiply scattering, and actively driven fluorescent systems with confocal differential dynamic microscopy. Physical Review Letters. 108 (21), 218103 (2012).
- Giavazzi, F. et al. Viscoelasticity of nematic liquid crystals at a glance. Soft Matter. 10 (22), 3938-3949 (2014).
- Bayles, A. V., Squires, T. M., Helgeson, M. E. Dark-field differential dynamic microscopy. Soft Matter. 12 (8), 2440-2452 (2016).
- Wulstein, D. M., Regan, K. E., Robertson-Anderson, R. M., McGorty, R. Light-sheet microscopy with digital Fourier analysis measures transport properties over large field-of-view. Optics Express. 24 (18), 20881-20894 (2016).
- Richards, J. A., Martinez, V. A., Arlt, J. Particle sizing for flowing colloidal suspensions using flow-differential dynamic microscopy. Soft Matter. 17 (14), 3945-3953 (2021).
- Ferri, F. et al. Kinetics of colloidal fractal aggregation by differential dynamic microscopy. The European Physical Journal Special Topics. 199 (1), 139-148 (2011).
- Lanfranco, R. et al. Adaptable DNA interactions regulate surface triggered self assembly. Nanoscale. 12 (36), 18616-18620 (2020).
- Cho, J. H., Cerbino, R., Bischofberger, I. Emergence of multiscale dynamics in colloidal gels. Physical Review Letters. 124 (8), 088005 (2020).
- Giavazzi, F., Trappe, V., Cerbino, R. Multiple dynamic regimes in a coarsening foam. Journal of Physics: Condensed Matter. 33 (2), 024002 (2020).
- He, K. et al. Diffusive dynamics of nanoparticles in arrays of nanoposts. ACS Nano. 7 (6), 5122-5130 (2013).
- Jacob, J. D. C., He, K., Retterer, S. T., Krishnamoorti, R., Conrad, J. C. Diffusive dynamics of nanoparticles in ultra-confined media. Soft Matter. 11 (38), 7515-7524 (2015).
- Sentjabrskaja, T. et al. Anomalous dynamics of intruders in a crowded environment of mobile obstacles. Nature Communications. 7, 11133 (2016).
- Hitimana, E., Roopnarine, B. K., Morozova, S. Diffusive dynamics of charged nanoparticles in convex lens-induced confinement. Soft Matter. 18 (4), 832-840 (2022).
- Wilson, L. G. et al. Differential dynamic microscopy of bacterial motility. Physical Review Letters. 106 (1), 018101 (2011).
- Martinez, V. A. et al. Differential dynamic microscopy: a high-throughput method for characterizing the motility of microorganisms. Biophysical Journal. 103 (8), 1637-1647 (2012).
- Germain, D., Leocmach, M., Gibaud, T. Differential dynamic microscopy to characterize Brownian motion and bacteria motility. American Journal of Physics. 84 (3), 202-210 (2016).
- Croze, O. A. et al. Helical and oscillatory microswimmer motility statistics from differential dynamic microscopy. New Journal of Physics. 21 (6), 063012 (2019).
- Safari, M. S., Vorontsova, M. A., Poling-Skutvik, R., Vekilov, P. G., Conrad, J. C. Differential dynamic microscopy of weakly scattering and polydisperse protein-rich clusters. Physical Review E. 92 (4), 042712 (2015).
- Wang, J., McGorty, R. Measuring capillary wave dynamics using differential dynamic microscopy. Soft Matter. 15 (37), 7412-7419 (2019).
- Cerbino, R., Giavazzi, F., Helgeson, M. E. Differential dynamic microscopy for the characterization of polymer systems. Journal of Polymer Science. 60 (7), 1079-1089 (2021).
- Cerbino, R., Cicuta, P. Perspective: differential dynamic microscopy extracts multi-scale activity in complex fluids and biological systems. The Journal of Chemical Physics. 147 (11), 110901 (2017).
- Drechsler, M., Giavazzi, F., Cerbino, R., Palacios, I. M. Active diffusion and advection in Drosophila oocytes result from the interplay of actin and microtubules. Nature Communications. 8 (1), 1-11 (2017).
- Burla, F., Sentjabrskaja, T., Pletikapic, G., Beugen, J. v., H. Koenderink, G. Particle diffusion in extracellular hydrogels. Soft Matter. 16 (5), 1366-1376 (2020).
- Regan, K., Wulstein, D., Rasmussen, H., McGorty, R., Robertson-Anderson, R. M. Bridging the spatiotemporal scales of macromolecular transport in crowded biomimetic systems. Soft Matter. 15 (6), 1200-1209 (2019).
- Wulstein, D. M., Regan, K. E., Garamella, J., McGorty, R. J., Robertson-Anderson, R. M. Topology-dependent anomalous dynamics of ring and linear DNA are sensitive to cytoskeleton crosslinking. Science Advances. 5 (12), eaay5912 (2019).
- Lee, G. et al. Active cytoskeletal composites display emergent tunable contractility and restructuring. Soft Matter. 17 (47), 10765-10776 (2021).
- Achiriloaie, D. H. et al. Kinesin and myosin motors compete to drive rich multi-phase dynamics in programmable cytoskeletal composites. arXiv:2112.11260 .(2021).
- Chen, X. et al. Coaxial differential dynamic microscopy for measurement of Brownian motion in weak optical field. Optics Express. 26 (24), 32083-32090 (2018).
- Reufer, M., Martinez, V. A., Schurtenberger, P., Poon, W. C. K. Differential dynamic microscopy for anisotropic colloidal dynamics. Langmuir. 28 (10), 4618-4624 (2012).
- Giavazzi, F., Haro-Pérez, C., Cerbino, R. Simultaneous characterization of rotational and translational diffusion of optically anisotropic particles by optical microscopy. Journal of Physics: Condensed Matter. 28 (19), 195201 (2016).
- Cerbino, R., Piotti, D., Buscaglia, M., Giavazzi, F. Dark field differential dynamic microscopy enables accurate characterization of the roto-translational dynamics of bacteria and colloidal clusters. Journal of Physics: Condensed Matter. 30 (2), 025901 (2017).
- Safari, M. S., Poling-Skutvik, R., Vekilov, P. G., Conrad, J. C. Differential dynamic microscopy of bidisperse colloidal suspensions. npj Microgravity. 3 (1), 21 (2017).
- Giavazzi, F., Pal, A., Cerbino, R. Probing roto-translational diffusion of small anisotropic colloidal particles with a bright-field microscope. The European Physical Journal E. 44 (4), 61 (2021).
- Schwarz-Linek, J. et al. Escherichia coli as a model active colloid: a practical introduction. Colloids and Surfaces B: Biointerfaces. 137, 2-16 (2016).
- Kurzthaler, C. et al. Probing the spatiotemporal dynamics of catalytic Janus particles with single-particle tracking and differential dynamic microscopy. Physical Review Letters. 121 (7), 078001 (2018).
- Mandal, S., Kurzthaler, C., Franosch, T., Löwen, H. Crowding-enhanced diffusion: an exact theory for highly entangled self-propelled stiff filaments. Physical Review Letters. 125 (13), 138002 (2020).
- Norouzisadeh, M., Chraga, M., Cerchiari, G., Croccolo, F. The modern structurator: increased performance for calculating the structure function. The European Physical Journal E. 44 (12), 146 (2021).
- Gu, M., Luo, Y., He, Y., Helgeson, M. E., Valentine, M. T. Uncertainty quantification and estimation in differential dynamic microscopy. Physical Review E. 104 (3), 034610 (2021).
- Hoyer, S., Hamman, J. xarray: N-D labeled arrays and datasets in Python. Journal of Open Research Software. 5 (1), 10 (2017).
- Aufderhorst-Roberts, A., Koenderink, G. H. Stiffening and inelastic fluidization in vimentin intermediate filament networks. Soft Matter. 15 (36), 7127-7136 (2019).
- Guo, M. et al. The role of vimentin intermediate filaments in cortical and cytoplasmic mechanics. Biophysical Journal. 105 (7), 1562-1568 (2013).
- Lavenus, S. B., Tudor, S. M., Ullo, M. F., Vosatka, K. W., Logue, J. S. A flexible network of vimentin intermediate filaments promotes migration of amoeboid cancer cells through confined environments. Journal of Biological Chemistry. 295 (19), 6700-6709 (2020).
- Patteson, A. E. et al. Loss of vimentin enhances cell motility through small confining spaces. Small. 15 (50), 1903180 (2019).
- Lin, Y.-C. et al. Origins of elasticity in intermediate filament networks. Physical Review Letters. 104 (5), 058101 (2010).
- Pawelzyk, P., Mücke, N., Herrmann, H., Willenbacher, N. Attractive interactions among intermediate filaments determine network mechanics in vitro. PLOS ONE. 9 (4), e93194 (2014).
- Schepers, A. V. et al. Multiscale mechanics and temporal evolution of vimentin intermediate filament networks. Proceedings of the National Academy of Sciences. 118 (27), e2102026118 (2021).
- Wu, H. et al. Effect of divalent cations on the structure and mechanics of vimentin intermediate filaments. Biophysical Journal. 119 (1), 55-64 (2020).
- Dogterom, M., Koenderink, G. H. Actin-microtubule crosstalk in cell biology. Nature Reviews Molecular Cell Biology. 20 (1), 38-54 (2019).
- Giavazzi, F., Malinverno, C., Scita, G., Cerbino, R. Tracking-free determination of single-cell displacements and division rates in confluent monolayers. Frontiers in Physics. 6, 120 (2018).
- Cho, J. H. Multiscale Probing of Colloidal Gelation Dynamics., Massachusetts Institute of Technology (2018).
- Krall, A. H., Weitz, D. A. Internal dynamics and elasticity of fractal colloidal gels. Physical Review Letters. 80 (4), 778-781 (1998).
- Bayles, A. V., Squires, T. M., Helgeson, M. E. Probe microrheology without particle tracking by differential dynamic microscopy. Rheologica Acta. 56 (11), 863-869 (2017).
- Edera, P., Bergamini, D., Trappe, V., Giavazzi, F., Cerbino, R. Differential dynamic microscopy microrheology of soft materials: a tracking-free determination of the frequency-dependent loss and storage moduli. Physical Review Materials. 1 (7), 073804 (2017).
- Wang, B., Kuo, J., Bae, S. C., Granick, S. When Brownian diffusion is not Gaussian. Nature Materials. 11 (6), 481-485 (2012).
- soft-matter/trackpy: Trackpy v0.5.0. Zenodo (2021).
- Crocker, J. C., Grier, D. G. Methods of digital video microscopy for colloidal studies. Journal of Colloid and Interface Science. 179 (1), 298-310 (1996).
- Nixon-Luke, R., Arlt, J., Poon, W. C. K., Bryant, G., Martinez, V. A. Probing the dynamics of turbid colloidal suspensions using differential dynamic microscopy. Soft Matter. 18 (9), 1856-1867 (2022).

