Microarray Analysis for Saccharomyces cerevisiae
Summary
In this protocol, gene expression in yeast (Saccharomyces cerevisiae) is changed after exposure to oxidative stress induced by the addition of hydrogen peroxide (H2O2), an oxidizing agent.
Abstract
In this protocol, gene expression in yeast (Saccharomyces cerevisiae) is changed after exposure to oxidative stress induced by the addition of hydrogen peroxide (H2O2), an oxidizing agent. In the experiment, yeast is grown for 48 hours in 1/2X YPD broth containing 3X glucose. The culture is split into a control and treated group. The experiment culture is treated with 0.5 mM H2O2 in Hanks Buffered Saline (HBSS) for 1 hour. The control culture is treated with HBSS only. Total RNA is extracted from both cultures and is converted to a biotin-labeled cRNA product through a multistep process. The final synthesis product is taken back to the UVM Microarray Core Facility and hybridized to the Affymetrix yeast GeneChips. The resulting gene expression data are uploaded into bioinformatics data analysis software.
Protocol
1. Isolating Total RNA from Saccharomyces cerevisiae using Enzymatic Lysis
- Prepare an RNase-free work zone using RNase ZAP (Ambion).
- Prepare fresh working lyticase reagent by adding 1 ml of RNase-free water directly to the lyticase vial to make a final working solution of 10 U/ul. Vortex well and invert several times to insure complete mixing. This 10 units/μl solution is stable for 12 hours.
- Prepare fresh DNase I solution using the Qiagen DNase kit and store on ice.
- Label a 1.7 ml microcentrifuge tube with your identification information. Transfer 1.5 ml of the appropriate yeast culture into the tube.
- Centrifuge the tube at 5000 x g (about 3/4 full speed) for 2 minutes at room temperature. Carefully remove the supernatant (without disturbing pellet) using a micropipette and discard the supernatant. **** Repeat step 4 if pellet size is too small or if indicated by instructor.****
- Add 1000 μl DEPC water to the pellet, vortex, and centrifuge at 5000 x g for 2 minutes. Discard the supernatant. Remove as much of the liquid as possible from the yeast pellet.
- Add the following to the yeast pellet and vortex to mix.
SGbuffer 10 μl Lyticasesolution(10u/μl) 30 μl - Incubate for 30 min at room temperature.
- Gently swirl the tube every 10 minutes to generate spheroplasts. Spheroplasts must be handled gently. Examine yeast under the microscope to observe complete spheroplasting. While examining the yeast on the microscope slide, add a small amount of 0.1% SDS to cause the yeast to swell and form perfect spheres (even for budding cells). This indicates that the cell walls have been digested.
- Add 350 μl b-RLT buffer to the tube and vortex vigorously for 1 minute. Ensure your tube is tightly capped by holding the lid closed while vortexing. This procedure will lyse the spheroplasts.
- Add 250 μl 100% ethanol to the tube and briefly vortex. A precipitate may form after the addition of ethanol, but this will not affect the collection of RNA.
- Collecting the RNA: Label an RNeasy spin column with your identification informationand carefully transfer all of the solution from step 10 to the spin column using a micropipette. Be careful not to touch the silica membrane with the pipet tip. Close the tube and centrifuge for 15 seconds at full speed. The RNA in the sample will adhere to the silica membrane of the spin column.
- Remove the spin column from the collection tube. Pipet the pass through liquid (the liquid in the collection tube) back onto the spin column and centrifuge again. Discard the collection tube (with the flow through fluid) and place the spin column into a new 2 ml collection tube.
- Add 350 μl RW1 buffer to the spin column. This solution is used to wash the RNA and remove salts and dissolved cellular debris. Close the tube and centrifuge for 15 seconds at full speed.
- Digesting the DNA: Remove the spin column from the centrifuge, open the tube lid and add 80 μl of DNase I solution to the middle of the spin column membrane. This step will digest any gDNA in the sample. Close the column lid and incubate at room temperature for 15 minutes.
- Transfer the spin column to a new 2 ml collection tube.
- Cleaning and washing the RNA: Add 350 μl RW1 buffer to the spin column and centrifuge at full speed for 15 seconds.
- Discard the collection tube and place the spin column into a new 2 ml collection tube.
- Add 500 μl RPE buffer to the spin column to wash the RNA on the silica membrane. Close the tube and centrifuge for 15 seconds at full speed.
- Discard the collection tube and place the spin column into a new 2 ml collection tube.
- Wash the silica membrane again by adding another 500 μl RPE buffer to the spin column. Close the tube and centrifuge for 15 seconds at full speed.
- Place the spin column in a new 2 ml collection tube and centrifuge in a microcentrifuge at full speed for 1 minute to ensure any residual liquid is removed from the silica membrane.
- Recovering the RNA: Transfer the RNeasy spin column to a new 1.7 ml microcentrifuge tube that has been labeled with your sample information. Note that the cap of the new tube will be left open for the next few steps.
- Carefully pipet 30 μl RNase-free water directly onto the center of the silica membrane. Do not touch the silica membrane with the pipet tip (see diagram). Look closely as you perform this step. Use both hands to guide the pipet. Make sure the water is evenly distributed on the membrane.
- Incubate the spin column at room temperature for 1 minute. Centrifuge at full speed for 30 seconds.
- Carefully transfer the 30 μl that is recovered in the 1.7 ml tube back onto the center of the silica membrane of the same spin column as in step 22 above. Place the column back in the same 1.7 ml collection tube and incubate for 1 minute at room temperature. Centrifuge for 1 minute at full speed. This double elution insures that the maximal amount of RNA is recovered from the membrane.
- Label two new 1.7 ml microcentrifuge tubes with your identification information and transfer all of the recovered RNA sample to one of these tubes.
- Transfer 4 μl of this sample to the other labeled tube. This sample will be transported back to UVM for Nanodrop quantification and RNA qualitative assessment (This is to validate RNA quantitative and qualitative analysis performed on site at participating institute).
- Place all samples on ice.
- RNA Quantification: Using the Eppendorf Biophotometer, measure the absorbance of the RNA sample on the spectrophotometer at a 1 to 50 dilution (1μl sample + 49μl H2O).
- Evaluate the RNA quality using the E-Gel (Invitrogen) 1.2% precast agarose gel for electrophoresis.
- RNA Qualitative Analysis: Set up the E-gel electrophoresis apparatus. Pre-run the gel for 2 minutes according to the manufacturer’s procedure. After the pre-run, remove the gel comb and add 14 μl of water to each well followed by 1μl of each sample to each well. Mix well by pipetting up and down. Use an appropriate size DNA Ladder in one well for size comparison later (BioLine Easy Ladder I size standard or Novagen PCR marker). Record which sample is in each lane. Run the gel for 20 minutes.
- Take a photo of the gel.
2. First Strand cDNA Synthesis
- Label a 0.5 ml PCR tube with your identification information. From the concentration of RNA determined from the above experiment, calculate the volume of the sample to obtain 3.0 ug. Transfer this amount to the tube. Add enough RNase-free water to bring the volume to 11.0 ul.
- Add 1.0 μl T7 oligo (dT)24 reagent to the tube. Make sure to pay particular attention to the pipet tip volume during this step to ensure that all the reagent was transferred. Vortex the tube and centrifuge at full speed for 5 seconds. Place the tube in a thermocycler set at 70°C for 10 minutes.
- While the 70°C incubation is in progress, prepare the first strand master mix in a new tube. Be sure to add the following reagents IN ORDER, vortex and centrifuge at full speed for 5 second and place the tube on ice.
First strand master mix: FirstStrandBuffer5X 4 μl 0.1MDTT 2 μl 10mMdNTP 1 μl SuperscriptII 1 μl
Use a P2 micropipette to measure volumes of 2 μl or less. - After the 70°C incubation in step 2 is complete, add 8 μl of the first strand master mix made in step 3 to the tube and incubate in a thermocycler at 42°C for 60 minutes. When first strand synthesis incubation is finished, place the tube on ice. During this incubation, prepare the second strand master mix.
3. Second Strand cDNA Synthesis
- Make the following second strand master mix in a new tube. Keep it on ice. Vortex and centrifuge all reagents before using. Add the reagents in the order listed.
Second strand master mix DEPC Water 91 ul 5X Second Strand Buffer 30 ul dNTPs (10mM) 3 ul Ecoli DNA Ligase (10U/ul) 1 ul Ecoli DNA Polymerase I (10U/ul) 4 ul Ecoli RNase H (2U/ul) 1 ul - Vortex the tube and centrifuge for 5 seconds at full speed.
- When the 42°C incubation is finished, transfer all 130 μl of the second strand master mix to the sample tube containing the RNA and first strand reagents. Vortex and centrifuge for 5 seconds at room temperature. Incubate the tube for 2 hours at 16°C in a thermocycler.
- At the end of the 2 hour incubation and while the sample is still at 16°C, add 2μl T4 DNA polymerase reagent to the tube. Briefly vortex and centrifuge the tube and incubate at 16°C for NO MORE THAN 5 additional minutes. Incubating longer then 5 minutes may reduce the quality of the cDNA due to the 3’to 5’exonuclease activity of the T4 polymerase.
- At the end of the 5 minute incubation, add 10 μl 0.5 M EDTA to stop the T4 DNA polymerase reaction. Store the tube at -20°C.
4. Precipitating the cDNA
- Centrifuge a Phase Lock Gel tube at full speed for one minute to insure the gel is at the bottom of the tube. DO NOT VORTEX PHASE LOCK TUBES.
- Add 162 μl of the bottom layer from the pH 8.0-Tris buffered Phenol/Chloroform/Isoamyl Alcohol (PCI) to the contents of the second strand cDNA synthesis reaction tube and vortex for 5 seconds to mix the contents (The total volume of the cDNA synthesis step from the above experiment is 162 ul. The PCI step requires equal volumes of aqueous and organic mixtures).
- Transfer all of the cDNA-PCI mixture to the phase lock gel tube using a micropipet. DO NOT VORTEX the phase lock gel tube.
- Centrifuge at full speed for 2 minutes.
- Label a new 1.7 ml microcentrifuge tube. Use a micropipet to transfer the top layer from the phase lock gel tube to the newly labeled 1.7 ml tube. Try to collect as much of the layer as possible.
- Add the following to the 1.7 ml microcentrifuge tube and vortex.
Ethanol (100%) 405 ul NH4OAc 80 ul Pellet Paint 1 ul - Place the tube in the centrifuge with the hinge of the tube facing out and centrifuge at full speed for 20 minutes at room temperature.
- GENTLY remove the tube from the centrifuge being careful not to disturb the cDNA pellet. The pellet should be pink and approximately the size of a grain of salt and on the side of the tube under the hinge. Place on ice and immediately proceed to next step.
5. Cleaning the cDNA Pellet
- Using a P1000 micropipet, carefully remove all of the supernatant from the tube. Be careful not to disturb the pellet. Remember that the pellet is your sample!
- Add 500μl cold 80% ethanol (stored in -20°C freezer) to the tube. Gently cap the tube and invert it slowly several times. Watch your pellet very closely. If the pellet becomes detached, place the tube back in rack and let the pellet settle to the bottom. Alternately, you may centrifuge the tube at full speed for 15 seconds to get the pellet back down to the bottom of the tube.
- Using a P1000 micropipet, carefully remove the ethanol being very careful not to disturb the pellet. Tip the tube to enable removal of as much liquid as possible.
- Repeat steps 2 and 3 with a new aliquot of 80% ethanol.
- Finally, remove all of the ethanol possible by using a P1000 micropipet. Centrifuge the tube at full speed for 5 seconds and using a P20 micropipet, remove the last few microliters of ethanol. The goal is to remove as much ethanol as possible without disturbing the pellet.
- Place the open tube in a rack or drying box for 10-20 minutes to evaporate the remaining ethanol. The dried pellet is easily lost once it is dry. Becareful to handle the tube gently. When done, close the cap. Visualize the dried pellet to confirm it is present in the tube.
- Resuspend the pellet in 22 μl of RNase-free water and place the tube on ice.
6. InVitroTranscription (IVT)
- The ENZO bioarray kit contains all the reagents needed to prepare biotin labeled cRNA from cDNA.
A master mixfor the entire class will be prepared as follows.The instructor will prepare this mix or designate someone from the class. This must be done in an RNase-free area.
Amt/sample #Samples Total Reagent 1 [10x Reaction buffer] 4μl Reagent 2 [10x Biotinnucleotides] 4μl Reagent 3 [10x DTT] 4μl Reagent 4 [10x RNase Inhibitor] 4μl Reagent 5 [20x T7 RNA polymerase] 2μl Total Volume 18 μl
NOTE: Prepare sufficient master mix for the number of samples plus one. This will insure enough for pipetting purposes. - Label a 0.5 ml PCR tube. Combine the following in the tube and pipet up and down several times to mix. Spin in a centrifuge at full speed for 5 seconds to get all reagents to the bottom of the tube.
cDNA mixture 22μl Enzomastermix[fromabove] 18μl TotalVolume 40μl - Incubate the tube at 37°C for16 hours in the thermocycler. When the reaction is complete, store the sample at -20°C
7. Cleaning the Biotinylated cRNA
- Transfer the entire cRNA sample to a new 1.7 ml microcentrifuge tube. Add 60 μl of RNase-free water and 350 μl RLT buffer and vortex for 5 seconds.
- Add 250 μl 100% ethanol and vortex again.
- Label an RNeasy spin column and transfer the entire cRNA sample to the column. Be careful not to touch the tip of the pipet to the silica membrane. Close the lid and centrifuge for 15 seconds at full speed.
- Remove the spin column from the collection tube. Pipet the pass through liquid (the liquid in the collection tube) back onto the spin column and centrifuge again for 15 seconds.
- Place the spin column into a new 2 ml collection tube and pipet 500 μl of RPE buffer onto the spin column. Close the tube and centrifuge for 15 seconds at full speed. Discard collection tube and the pass through liquid. Place the spin column into a new 2 ml collection tube.
- Add another 500 μl RPE buffer to the spin column.
- Transfer the spin column into a new collection tube and perform a column “drying” spin at full speed for 2 minutes.
- Label a new 1.7 ml microcentrifuge tube with your identification information. Transfer the spin column to this tube.
- Pipet 30 μl RNase-free water onto the center RNeasy silica membrane and incubate at room temp for 1 minute. Close the tube and centrifuge for 1 minute at full speed.
- Carefully transfer the 30 μl that is recovered in the 1.7 ml tube back onto the center of the RNeasy silica membrane of the same spin column. Place the column back in to the same 1.7 ml collection tube and incubate for 1 minute at room temperature. Centrifuge for 1 minute at full speed. This double elution insures that the maximal amount of cRNA is recovered from the membrane.
- Transfer this clean biotin-labeled cRNA to a new 1.7 ml centrifuge tube and label appropriately.
- Determine the cRNA concentration using the same procedure as outlined above during the RNA isolation procedure. Record the concentration and 260/280 ratio.
8. Fragmenting the cRNA for Target Preparation
- Label a 0.5 ml PCR tube with your identification information. Transfer 5 ug of the equivalent amount of cRNA to the tube. Add enough RNase-free water to bring the total volume to 16.0 ul, and then add 4.0 μl of 5X fragmentation buffer to the tube. The total volume in the tube should be 20.0 ul.
- Vortex the tube and centrifuge for 10 seconds. Incubate the tube at 94°C for 30 minutes in a thermocycler. Put on ice following the incubation.
9. Assessing the Fragmented and Unfragmented cRNA using Agarose Gel
- Set up the E-gel electrophoresis apparatus using a 1.2 % precast agarose gel. Pre-run the gel for 2 minutes according to the manufacture’s recommendation
- Add 14 μl of water to each well on the E-gel.
- Add 2μl of each sample to each well and pipet up and down to mix well. Analyze both the fragmented and unfragmented cRNA of each sample. It is best to load the samples (fragmented and unfragmented) in neighboring lanes. Record the identity of each sample in each lane.
- Add 4 μl of a DNA ladder (BioLine Easy Ladder I size standard or Novagen PCR marker) to one lane of the gel
- Run the gel for 20 minutes.
- Visualize the E-gel on a transilluminator. Take a gel picture.
10. Hybridization to the Yeast 2.0 GeneChip
Representative Results:
Figure 1. A scanned Affymetrix Yeast GeneChip Image (Affymetrix GeneChip Operating Software (GCOS))
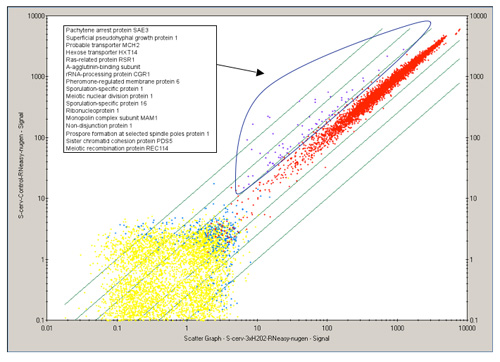
Figure2. 2D scatter plot of all genetic transcripts (~6,700 genes), comparing a control and treated yeast data. Each point represents a single gene. Genes colored in purple indicate genes that are differentially expressed while genes colored in red are not. Descriptions for the differentially expressed genes are labeled in the corresponding legend and most are involved in cell cycle control in this example. (Affymetrix GeneChip Operating Software (GCOS))
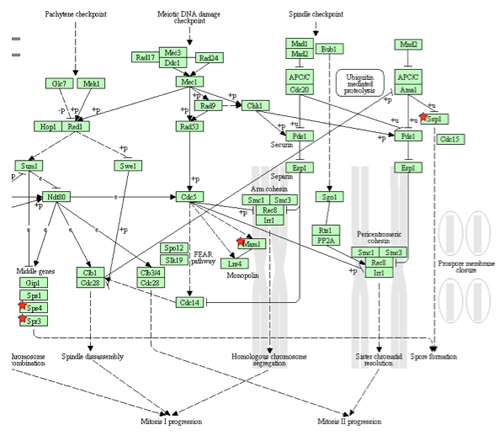
Figure 3. This flowchart illustrates differentially expressed genes in an affected biological pathway. The genes indicated with a red star indicate the down-regulated genes in the meiotic pathway. (The Database for Annotation, Visualization and Integrated Discovery (DAVID)
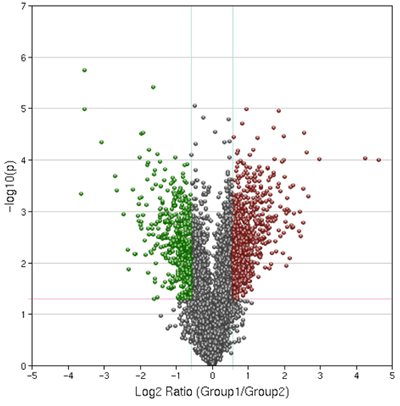
Figure 4. Representative Results of a Volcano Plot. Control and treated samples were compared with a p-value cut off of .05 and a 1.5 fold expression change cut off. This plot was generated using Geospiza’s Genesifter software kindly donated to the students for educational purposes.
Discussion
Applications and significance.
The Vermont Genetics Network outreach program, at the University of Vermont, conducts undergraduate outreach to eight partner baccalaureate colleges throughout the state. The goal of the VGN Outreach Core is to expose undergraduates in the state of Vermont to state-of-the-art scientific technology & resources using hands-on experiences. The microarray module described was developed in 2003 and subsequently upgraded. It has been offered as a mini-course or integrated into existing laboratory curriculum at the participating institutions.
This project, between research university cores and undergraduate colleges, promotes collaborations in education and research. Microarray outreach throughout the state has enhanced curriculum and created research and networking opportunities for core facilities, faculty and students.
As undergraduate faculty adopt the program they are encouraged to design unique experiments provided there are appropriate Affymetrix GeneChips and annotation available. All information for this module including the laboratory manual, references and PowerPoint presentations are available online (Vermont Genetics Network, 2008).
Critical steps:
Spheroplasting: It is important to observe successful spheroplasting under the microscope. The use of SDS is highly beneficial as it causes the yeast swelling resulting in spheroids. It is important to observe that the budding cells form spheroids as well. If partial spheroplasting is observed, an extended treatment with lyticase is recommended.
Pellet cleaning: During the precipitation reaction and subsequent ethanol washing step, it is very easy to lose the cDNA pellet. Extreme care and meticulous attention to the pellet is imperative.
Applying the water to the center of the membrane: During the RNA elution step from the membrane, it is important to “squirt” the 30 μl of water directly to the center of the silica membrane. If this is not accomplished, the column can be centrifuged and the resulting elutant can be re-applied to the silica membrane again. This can be repeated as many times as necessary.
Modifications and Product Substitutions:
- Alternative RNA clean-up columns can be substituted. Examples are the USB prep-ease and the Invitrogen Pure Link RNA kit, and the Omega RNA clean-up kit to name a few.
- Schizosaccharomyces pombe is an optional yeast. The Affymetrix GeneChip contains both genomes on the same chip
- Various treatments and times may be used. This may include drug treatments, temperature treatments, anti-oxidants, etc. Treatment times may also be adjusted.
- DNase treatment may not be required because the methods described employ the use of an Oligo (dT) primer.
- It is not required to perform a double elution of the RNA off the column with the same 30 μl aliquot. Once is almost always adequate. This is suggested to insure that a complete recovery is obtained. Additionally, the water used to elute the RNA from the column can be pre-heated to 65°C to further enhance the ability to recover RNA from the silica column.
- Many methods are available to quantify RNA. The Eppendorf and Nanodrop have only been selected for their ease of use and tested accuracy.
- Agarose gel electrophoresis many be performed using standard laboratory equipment. The E-Gel is not required but is desirable because it is RNase free and does not require gel solidification waiting time.
- Ordering information for products has been supplied. However, many general reagents can be ordered from multiple vendors.
- One Cycle cDNA reagents can be purchased separately if that is more cost effective.
- There are other target prep methods but we feel this one offers more teaching opportunities for the student.
Disclosures
The authors have nothing to disclose.
Acknowledgements
University of Vermont, Vermont Genetics Network. This publication was made possible by the Vermont Genetics Network, through Grant Number P20 RR16462 from the BRIN Program and Grant Number P20 RR16462 from the INBRE Program of theNational Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
We would like to thank all our participating outreach institutions for their collaboration and input in refining this module Castleton State College, Green Mountain College, Johnson State College, Lyndon State College, Marlboro College, Middlebury College, Norwich University and Saint Michael’s College.
Materials
| General Laboratory Equipment | Company | Product number |
| Spetrophotometer | ||
| Thermocyler | ||
| Microcentrifuge | ||
| Vortex | ||
| Stir Plates (2) | ||
| P-10s pipettors | ||
| P-20’s pipettors: | ||
| P-200’s pipettors: | ||
| P-1000’s pipettors: | ||
| Camera and transilluminator | ||
| Supplies – recommended | ||
| E-Gel Apparatus | Invitrogen | G6000-08 |
| E-Gels | Invitrogen | G6018-02 |
| Axygen 1.7 ml. Clear | Krackler | 383-MCT175C |
| Axygen 0.5 ml clear tubes | Krackler | 383-MCT060C |
| Axygen 2ml capture tubes | Krackler | 383-MCT200NC |
| boxes of P-10 ART tips: | CLP | BT10XL |
| Boxes of P-100 ART tips: | CLP | BT200 |
| Boxes of P-20 ART Tips: | CLP | BT20 |
| Boxes of P-1000 ART Tips: | CLP | BT1000 |
| Yeast Chips | ||
| General Laboratory Supplies | ||
| 25 ml sterile disposable pipets- | ||
| 5ml sterile disposable pipets- | ||
| Microfuge tube racks | ||
| KimWipes: | ||
| Lab Coats | ||
| Stir Bars | ||
| Culture flasks | ||
| Innoculating loops | ||
| Sharpies | ||
| Gloves | ||
| Autoclave tape | ||
| Stirrer bars | ||
| Reagents | ||
| 30% Hydrogen Peroxide | EMD Millipore | 386790 |
| HBSS without Ca, Mg, or dye | Sigma-Aldrich | H6648-100ml |
| Qiagen Rneasy Mini Kit: | Qiagen | 74104 |
| Qiagen DNase Kit: | Qiagen | 79254 |
| Rnase Remover | Denville Scientific | D1180 |
| Harleco Alcohol 100% | EMD Millipore | 65347 |
| ENZO IVT Kit | ENZO | ENZ-42655-10 |
| Lyticase: one bottle | VWR | IC19012310 |
| Water, Cell Culture grade | EMD Millipore | 4.86505 |
| Yeast culture: | ATCC | 18824 |
| YPD broth powder | EMD Millipore | 4.8504 |
| D(+) Glucose, Anhydrous | EMD Millipore | 346351 |
| Sorbitol | EMD Millipore | 56755 |
| EDTA | EMD Millipore | 4005 |
| SDS solution, 20% | EMD Millipore | 7990-OP |
| Pellet Paint | EMD Millipore | 69049 |
| PCI RNase DNase free (7 aliquots): | Fisher | AC32711-500 |
| 7.5M NH4OAc | Fisher | ICN1987 5980 |
| Fragmentation Buffer (200 mM Tris-Acetate, pH 8.1, 500 mM KOAc, 150 mM MgOA) | ||
| Trizma Base | Fiaher | 50-899-90034 |
| KOAc | Fisher | NC9757725 |
| MgOA | Fisher | NC9681042 |
| Loading Dye | Invitrogen | 10482055 |
| PCR Markers | EMD Millipore | 69278-3 |
| Phase Lock Gels | Fisher | FP2302820 |
| One-Cycle cDNA Kit | Invitrogen | A10752-030 |
| Includes; | ||
| E. coli DNA Pol | ||
| dNTP’s | ||
| T4 DNA Pol. | ||
| T-7 oligod(T) | ||
| 0.5ml EDTA pH 8.0 | ||
| First Strand Buffer | ||
| E. Coli RNaseH | ||
| Second Strand Buffer | ||
| E. Coli DNA ligase | ||
| SuperScript II | ||
| DTT | ||
References
- Van Gelder, R. N., von Zastrow, M. E., Yool, A., Dement, W. C., Barchas, J. D., Eberwine, J. H. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A. 87, 1663-1667 (1990).
- Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D., Brown, P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 11, 4241-4257 (2000).
- D’Autréaux, B., Toledano, M. B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 8, 813-824 (2007).
- Shenton, D., Smirnova, J. B., Selley, J. N., Carroll, K., Hubbard, S. J., Pavitt, G. D., Ashe, M. P., Grant, C. M. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 281, 29011-29021 (2006).
- Godon, C., Lagniel, G., Lee, J., Buhler, J. M., Kieffer, S., Perrot, M., Boucherie, H., Toledano, M. B., Labarre, J. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 273, 22480-229 (1998).
- Vermont Genetics Network. . Microarray Outreach. , (2008).
- . . Affymetrix GeneChip Expression Analysis: Technical Manual. , (2004).
- Huang, D. W., Sherman, B. T., Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 4, 44-57 (2009).
- Dennis, G., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., Lempicki, R. A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3-P3 (2003).

