Label-Retention Expansion Microscopy (LR-ExM) Enables Super-Resolution Imaging and High-Efficiency Labeling
Summary
A protocol of label retention expansion microscopy (LR-ExM) is demonstrated. LR-ExM uses a novel set of trifunctional anchors, which provides better labeling efficiency compared to previously introduced expansion microscopies.
Abstract
Expansion microscopy (ExM) is a sample preparation technique that can be combined with most light microscopy methods to increase the resolution. After embedding cells or tissues in swellable hydrogel, samples can be physically expanded three-to sixteen-fold (linear dimension) compared to the original size. Therefore, the effective resolution of any microscope is increased by the expansion factor. A major limitation of the previously introduced ExM is reduced fluorescence after polymerization and the digestion procedure. To overcome this limitation, label-retention expansion microscopy (LR-ExM) has been developed, which prevents signal loss and greatly enhances labeling efficiency using a set of novel trifunctional anchors. This technique allows one to achieve higher resolution when investigating cellular or subcellular structures at a nanometric scale with minimal fluorescent signal loss. LR-ExM can be used not only for immunofluorescence labeling, but also with self-labeling protein tags, such as SNAP- and CLIP-tags, thus achieving higher labeling efficiency. This work presents the procedure and troubleshooting for this immunostaining-based approach, as well as discussion of self-labeling tagging approaches of LR-ExM as an alternative.
Introduction
Expansion microscopy (ExM) has been used by researchers since it was first introduced as a convenient approach to achieve super resolution imaging with conventional microscopes, such as epifluorescence and confocal microscopes1,2,3,4,5,6,7. Using ExM, it is possible to achieve ~70 nm lateral resolution even with regular confocal microscopes. When ExM is combined with super-resolution imaging, the resolution is further improved. For instance, one can achieve roughly 30 nm resolution with structured illumination microscopy (SIM), and roughly 4 nm resolution with stochastic optical reconstruction microscopy (STORM)1,5.
However, low labeling efficiency is a critical issue with standard ExM methods. Fluorescence loss can vary based on the type of fluorescent groups and digestion time. On average, however, it has been reported that more than 50% of fluorophores are lost after the polymerization and the protein digestion steps of ExM, which is detrimental to the imaging quality3,4.
Thus, label-retention expansion microscopy (LR-ExM) has been developed, which can efficiently retain labels and reduce signal loss1. The key innovation of LR-ExM is the use of a set of trifunctional anchors instead of merely using fluorescent dyes-as in the standard ExM procedure-for staining proteins of interest. These trifunctional linkers consist of three parts: (1) the connector (e.g., N-hydroxysuccinimide (NHS)) to connect to the antibody, (2) the anchor (e.g., methacrylamide (MA)) to anchor proteins to the polymer, and (3) the reporter (e.g., biotin or digoxigenin (DIG)) to conjugate to an organic dye. The trifunctional anchors survive the polymerization and protein digestion steps, and therefore prevent fluorophore loss.
Furthermore, this method holds great potential since it is compatible with self-labeling enzymatic tags such as SNAP or CLIP. Enzymatic tag approaches have some benefits over the immunostaining approach regarding high specificity and labeling efficiency8,9,10.
In this manuscript, a detailed procedure of LR-ExM is demonstrated. LR-ExM is a highly effective and flexible method to achieve high spatial resolution with enhanced labeling efficiency.
Protocol
1. Cell culture
- Use U2OS cells cultured in McCoy's 5A medium supplemented with 10% FBS at 37 °C in 5% CO2.
- Culture cells onto a 16 well removable chambered coverglass (culture area 0.4 cm2) for ease of handling.
2. Fixation and permeabilization
NOTE: Fixation and permeabilization conditions depend on the optimized immunostaining protocols. The following is a fixation and permeabilization protocol to co-immunostain microtubule and clathrin coated pits (CCPs).
- Once cell counts reach ~0.04 x 106, fix cells with 100 µL of 3.2% paraformaldehyde (PFA) in PEM buffer (100 mM PIPES, 1 mM EGTA, and 1 mM MgCl2, pH 6.9) for 10 min at room temperature (Table 1).
NOTE: Fixation using PFA is performed in a certified safety hood. - Wash cells three times for 5 min each with 200 µL of phosphate-buffered saline (PBS).
- Permeabilize cells with the 100 µL of permeabilization buffer at room temperature for 15 min (Table 1).
NOTE: Proceed with labeling as soon as possible to avoid target protein, RNA or DNA degradation, and to obtain an optimal result. If it is needed, however, fixed samples can be stored for an elongated time (up to a month) in a PBS buffer at 4 °C. Replace any evaporated PBS and protect the sample from photobleaching.
3. Endogenous streptavidin and biotin blocking
- Incubate cells with the streptavidin solution diluted in a cell blocking buffer (100 µL) for 15 min at room temperature (Table 1).
- Rinse briefly with 200 µL of cell blocking buffer.
- Incubate cells with 100 µL of biotin solution diluted in a cell blocking buffer for 15 min at room temperature.
NOTE: When using a self-labeling tagging approach, such as using SNAP- or CLIP- tags, skip step 4, and proceed to step 5.1: self labeling tagging approach.
4. Primary antibody staining
- Incubate cells with rat anti-α-tubulin antibody (1:500 dilution) and rabbit anti-clathrin heavy-chain antibody (1:100 dilution) in 100 µL of cell blocking buffer for 16 h at 4 °C or 1 h at room temperature. Double the incubation time for the tissue samples.
- Wash samples four times with 200 µL of PBS for 5 min each.
5. LR-ExM specific secondary antibody staining
- Conjugate IgG antibodies with the trifunctional linkers such as N-hydroxysuccinimide (NHS) – methacrylamide (MA) -Biotin or NHS-MA-Digoxigenin (Dig) (Figure 1B). Synthesis of trifunctional anchors and conjugation of the secondary antibodies are previously introduced in Shi et al.1. Self-labeling tagging approach: Conjugate IgG antibodies with the trifunctional linkers, such as MA- benzylguanine (BG)-Biotin or MA- benzylcytosine (BC)-Dig (Figure 1B). Synthesis of trifunctional anchors and a conjugation of the secondary antibodies are previously introduced in Shi et al.1.
- Incubate cells with the donkey anti-rabbit-Dig-MA (1:100 dilution) and donkey anti-rat-biotin-MA (1:100 dilution) secondary antibodies in 100 µL of cell blocking buffer for 1 h at room temperature. Double this incubation time for the tissue samples.
- Wash samples four times in 200 µL of PBS for 5 min each.
6. Additional anchoring
- Incubate cells with 0.25% glutaraldehyde (GA) in PBS (100 µL) for 15 min at room temperature in order to anchor the proteins onto the hydrogel. Alternatively, use 25 mM methacrylic acid N-hydroxysuccinimide ester (MA-NHS) for anchoring. One hour incubation at room temperature is encouraged.
- Wash samples three times in PBS for 5 min each.
7. Gelation
NOTE: The expansion speed is determined by the diffusion time of salt and water out or into the gel; thus, casting thin gels speeds up expansion time.
- Remove the upper structure of the 16-well gel plate. Add 40 µL of monomer solution to each well to condition cells on ice. Incubate on ice for 5 min.
- To prepare a monomer solution, see Table 2.
- Prepare the gelation solution on ice. Add double distilled water and 10% N,N,N′,N′ Tetramethylethylenediamine (TEMED) accelerator to the monomer solution (10% TEMED: monomer solution = 1:47); do not add the initiator yet (Table 3).
- For a cell culture chamber with an area of 0.4 cm2, use a gelation solution volume of about 40 µL. Prepare 45 µL of gelation solution for each well.
- Add 10% ammonium persulfate (APS) to the gelation solution, incubate on ice, and immediately pipette 40 µL of gelation solution into each silicone gasket well on ice. Incubate on ice for 3 min (10% APS:10% TEMED:monomer solution = 1:1:47).
- Protect the sample from light and move the coverglass in the 10 cm Petri dish to a 37 °C incubator for 1.5 h for gelation. To keep the humidity of the gel during the 37 °C incubation, cover the Petri dish with the lid, and put a few drops of water in the Petri dish.
8. Digestion
- After gelation, remove the glass to separate the gel and transfer the gel to 6 well plate. Digest embedded cells with 2 mL of digestion buffer (Table 1) overnight at room temperature or 4 h at 37 °C. Use at least 10-fold excess volume of digestion buffer.
- Wash gels with at least a 10-fold excess volume of water than the final gel volume. Repeat the washing step four times for 20 to 30 min each time. The gel expands by about two-fold in each dimension.
NOTE: Gel samples can be stored for up to 1 month in a PBS buffer at 4 °C. Replace any evaporated water to avoid drying, and protect the sample from light during storage. To avoid degradation of trifunctional anchors, samples should be stained as soon as possible after digestion and wash.
9. Post-digestion fluorescence staining
- Incubate gels in 2 mL volume of streptavidin (STV)/digoxigenin (DIG) staining buffer with 2 to 5 µM STV-dye and/or anti-DIG-dye for 24 h at room temperature. Keep the samples in the dark.
10. Expansion
- Wash and expand the gel four times with excessive water (2-3 mL) for 30 min to 1 h each time.
- For easier visualization of cells under fluorescent microscopy, stain cells with DAPI during the third of the five washes. Dilute DAPI stock concentration (5 mg/mL, stored at 4 °C) in 1:5,000 dilutions in wash water. Incubate the gel with a DAPI-water solution (2 mL) for 30 min to 1 h.
- Wash two additional times with 3 mL of water.
- Expand gels in any flat dish that is big enough to contain the samples. The expanded gels which are prepared on 0.4 cm2 area coverglass fit nicely in a glass bottom 6-well plate.
NOTE: Post-expansion-stained samples can be stored for up to 4 months in a PBS buffer at 4° C. Add enough water to avoid drying and protect the sample from photobleaching. Repeat the expansion and DAPI staining step before imaging. To avoid any risk of fluorophore degradation, samples need to be imaged as soon as possible.
11. Imaging
- Coat the 6-well glass bottom imaging chamber with 0.01% poly-L-lysine (3 mL each) to immobilize the gel samples.
- Transfer gel samples to the coated imaging chamber.
- Image the samples with any desired fluorescence scopes.
Representative Results
Clathrin-coated pits (CCPs) are immunostained using trifunctional anchors (Figure 1B) and LR-ExM is performed as described in Figure 1A. LR-ExM (Figure 2C,E) shows much higher fluorescence intensity compared to the protein-retention expansion microscopy (proExM, Figure 2A) or biotin-ExM (Figure 2B); the signal for LR-ExM was about six times higher than proExM (Figure 2D). LR-ExM can effectively capture small structures such as CCPs, which are even smaller than the diffraction limit (Figure 2F,G).
Some representative results of LR-ExM are showcased by using the immunostaining approach. Microtubules and CCPs are co-immunostained using trifunctional anchors, including NHS-MA-DIG and NHS-MA-biotin (Figure 3A,B). LR-ExM is available for the enzymatic tag-based approach. The result is demonstrated using trifunctional anchors BG-MA-biotin (for SNAP-tag) and BC-MA-DIG (for CLIP-tag) in Figure 3C–F. Furthermore, one can combine both immunostaining and the protein-tag approach in LR-ExM as shown in Figure 3G–J. From these results, it is confirmed that LR-ExM is beneficial to obtain high quality images with enhanced labeling efficiency and resolution.
LR-ExM shows great performance in tissue samples as well. LR-ExM is performed on the mouse brain tissue by co-immunostaining the presynaptic marker Bassoon and postsynaptic marker Homer1. The two labels are clear and well separated, which support high resolution and labeling efficiency (Figure 4A–E).
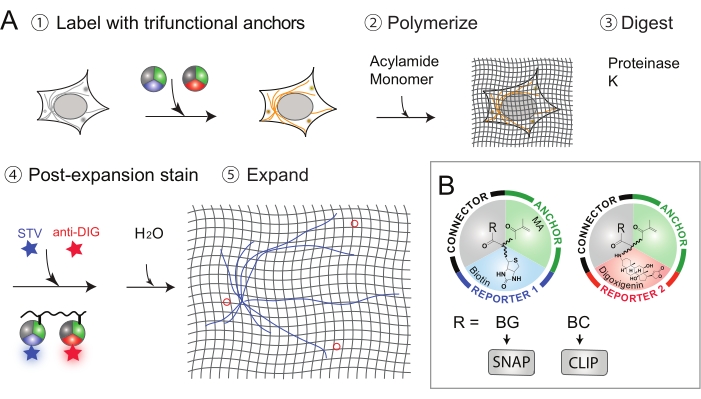
Figure 1: Workflow of Label Retention Expansion Microscopy (LR-ExM). (A) Workflow of LR-ExM. (B) Schematics of trifunctional anchors. This figure has been modified from Shi et al.1. Please click here to view a larger version of this figure.
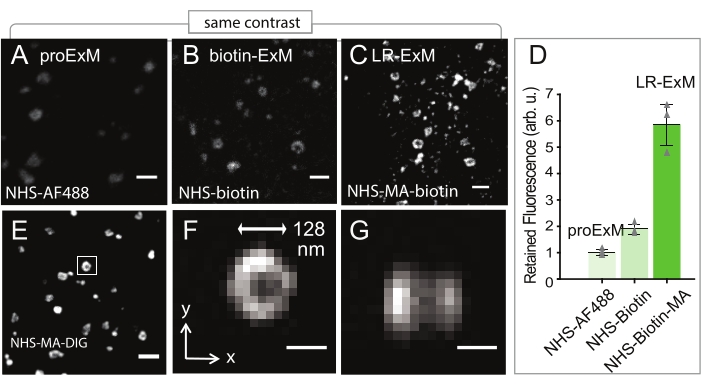
Figure 2: Quantification of signal retention (A–C) Expansion Microscopy (ExM) confocal images of clathrin-coated pits (CCPs) in U2OS cells indirectly immunostained for clathrin heavy-chain (POI). (A) Protein-retention ExM (ProExM) using AF488-conjugated secondary antibodies. (B) ExM with post-expansion labeling using biotin-conjugated antibodies. (C) LR-ExM using antibodies conjugated with NHS-MA-biotin trifunctional anchors. Samples in B and C are post-expansion stained with streptavidin-AF488. (D) Intensity quantification of A-C. Error bars represent SD. n = 3 for each case. The length expansion ratios for images in A, B, C, and E-G are 4.3, 4.5, 4.6, and 4.3, respectively. The length expansion ratio for the samples used in plot D is 4.5 ± 0.2. Scale bars, 500 nm (A-C and E), and 100 nm (F and G). All scale bars are in pre-expansion units. arb. u., arbitrary units; STV, streptavidin. All images are obtained by using a spinning-disk confocal microscope (Nikon CSU-W1) with a 60x water immersion objective (NA1.27). This figure has been modified from Shi et al.1. Please click here to view a larger version of this figure.
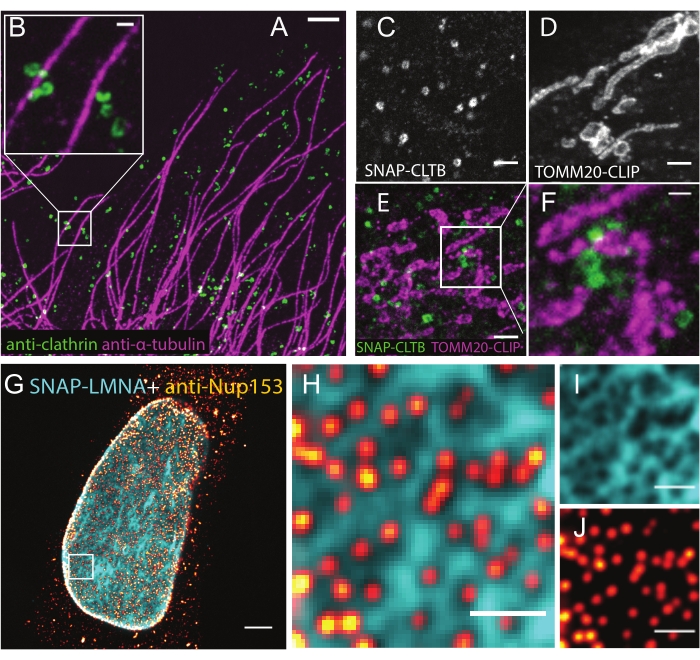
Figure 3: Representative results of LR-ExM using confocal microscopes. (A,B) LR-ExM confocal images of microtubules labeled with NHS-MA-biotin-conjugated secondary antibodies (magenta) and CCPs labeled with NHS-MA-DIG-conjugated secondary antibodies (green) in a U2OS cell. (B) Magnified view. (C-F) LR-ExM confocal images of CCPs and/or mitochondria in HeLa cells labeled using protein tag approach (C) SNAP tag-labeled clathrin, (D) CLIP tag-labeled TOMM20, and (E) two-color imaging. (F) Magnified view. (G) Two-color confocal LR-ExM images using a combination of immunostaining approach (NPC, red hot) and protein-tag approach (SNAP-tagged lamin A/C (cyan)). (H) Magnified view. (I,J) Views of individual channels of H. Note the cytoplasmic background in G is caused by the anti-NUP153 antibody. The length expansion ratios for images in A-B: 4.7, C-D: 4.4, E-F: 4.5, and G-J is 4.5. Scale bars,1 µm for A, 200 nm for B and F, 500 nm for C-E, 2 µm for G, and 500 nm for H, I, and J.All scale bars are in pre-expansion units. All the images are obtained by using a spinning-disk confocal microscope (Nikon CSU-W1) with a 60x water immersion objective (NA1.27). This figure has been modified from Shi et al.1. Please click here to view a larger version of this figure.
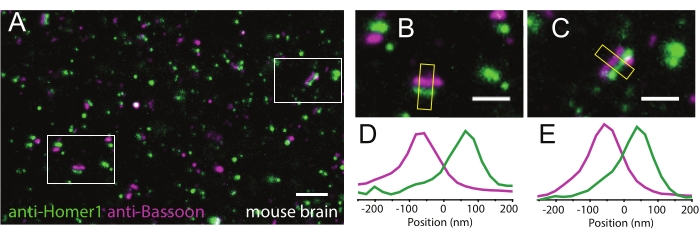
Figure 4: Representative results of LR-ExM on tissue samples. (A) LR-ExM confocal image of mouse brain slice indirectly immunostained for the presynaptic marker Bassoon (magenta) and the postsynaptic marker Homer1 (green). (B,C) Zoomed-in images of synapses. (D,E) Transverse intensity profiles along the yellow box long axes. Bassoon is labeled with NHS-MA-DIG-conjugated secondary antibodies, and Homer1 is labeled with NHS-MA-biotin-conjugated secondary antibodies. All samples are post-expansion stained with streptavindin-AF488 and or anti-Digoxin-AF594. The length expansion ratios for images in A-C are 4.2. Scale bars, 1 µm (A), and 200 nm (B,C). All scale bars are in pre-expansion units. All the images are obtained by using a spinning-disk confocal microscope (Nikon CSU-W1) with a 60x water immersion objective (NA1.27). This figure has been modified from Shi et al.1. Please click here to view a larger version of this figure.
Table 1: Chemicals and buffers Please click here to download this Table.
Table 2: Monomer solution Please click here to download this Table.
Table 3: Gelation solution Please click here to download this Table.
Discussion
The key innovation of LR-ExM is to use trifunctional anchors to effectively label the target proteins and improve image quality. This method is limited by trifunctional anchors, which are not so readily available to researchers. However, trifunctional anchors can be shared with other researchers upon request, and similar products such as ExM probes from Chrometa are now commercially available as well.
In this protocol, 1 h incubation at room temperature has been performed for the primary and secondary antibody staining steps. However, these steps can alternatively be done overnight at 4 °C, and it is observed that overnight staining lowers the background noise.
In order to prevent structural distortion derived from anisotropic expansion of the hydrogel, it is important to perform a thorough protein digestion using protease3. However, previous expansion microscopies show more than 50% of fluorescence labels can be lost after polymerization and protein digestion steps, which results in poor image quality3,4. In order to resolve this issue, LR-ExM has been developed, which minimizes the loss of signal by introducing fluorescent labels after digestion1.
LR-ExM can be widely used with immunostaining-based methods to study the target structure. Furthermore, LR-ExM can be combined with self-labeling protein tag approaches such as SNAP- or CLIP-tag based approaches. This is helpful especially when good antibodies of the interest are not available or if one needs improved target specificity.
In this paper, the protocol of LR-ExM with the length expansion factor of around 4 is introduced. However, LR-ExM is not limited to certain expansion ratios or types of samples. In fact, this method can be combined with any pre-existing expansion microscopies, such as X10 expansion microscopy4,11 or TREx12 to achieve higher resolution. LR-ExM can also be combined with pan-ExM, so that it effectively visualizes the whole protein landscape with high resolution without sacrificing specificity13.
In summary, LR-ExM has the potential to be widely used by many researchers as a tool to elucidate cellular structures with high resolution and labeling efficiency.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the U.S. National Institutes of Health (R00 GM126136 to X.S.), the U.S. National Science Foundation (DMS1763272 to S.P.) and the Simons Foundation (594598 to S.P.).
Materials
| Acrylamide | Sigma | A9099 | ExM Gel |
| AffiniPure Donkey Anti-Rabbit IgG | Jackson ImmunoResearch | H+L, 711–005-152 | Antibody |
| AffiniPure Donkey Anti-Rat IgG | Jackson ImmunoResearch | H+L, 712–005-153 | Antibody |
| Alexa Fluor 488-Streptavidin | Jackson ImmunoResearch | 016-540-084 | Fluorescent probes |
| Alexa Fluor 594 Streptavidin | Jackson ImmunoResearch | 016-580-084 | Fluorescent probes |
| Alexa Fluor 647 Streptavidin | Jackson ImmunoResearch | 016-600-084 | Fluorescent probes |
| Ammonium Persulfate | Sigma | A3678 | ExM Gel |
| anti-H3K4me3 | Abcam | ab8580 | Antibody |
| anti-H3K9me3 | Abcam | ab176916 | Antibody |
| DAPI dilacetate | Thermofisher Scientific | D3571 | Fluorescent probes |
| DyLight 488 Labeled Anti-Digoxigenin/Digoxin (DIG) | Vector Laboratories | DI-7488 | Fluorescent probes |
| DyLight 594 Labeled Anti-Digoxigenin/Digoxin (DIG) | Vector Laboratories | DI-7594 | Fluorescent probes |
| EGTA | EMD Millipore Corp. | 324626-25GM | Fixation buffer |
| Ethylenediaminetetraacetic acid | Sigma | EDTA | Digestion buffer |
| Glutaraldehyde 10% EM Grade | Electron Microscopy Sciences | 50-262-13 | Anchoring |
| Grace Bio-Labs CultureWell removable chambered coverglass | Grace Bio-Labs | GBL112358-8EA | Cell culture chamber |
| Grace Bio-Labs CultureWell removal tool | Grace Bio-Labs | GBL103259 | Removal tool |
| Guanidine HCl | Sigma | G3272 | Digestion buffer |
| Magnesium chloride | Sigma | M8266-1KG | Fixation buffer |
| McCoy's 5a | ATCC | 30–2007 | Celll culture medium |
| Methacrylic acid N-hydroxysuccinimide ester,98% (MA-NHS) | Sigma | 730300-1G | Anchoring |
| monoclonal mouse anti-Nup153 antibody | Abcam | ab24700 | Antibody |
| N,N′Methylenebisacrylamide | Sigma | M7279 | ExM Gel |
| N,N,N′,N′ Tetramethylethylenediamine (TEMED) | Sigma | T7024 | ExM Gel |
| 16% Paraformaldehyde Aqueous Solutions | Electron Microscopy Sciences | 50-980-487 | Fixation buffer |
| PIPES | Sigma | P6757-25G | Fixation buffer |
| Poly-L-Lysine | Sigma | P8920-100ML | Chamber coating |
| Proteinase K | Sigma-Aldrich | P4850-5ML | Digestion buffer |
| Rabbit anti-clathrin heavy-chain antibody | Abcam | ab21679 | Antibody |
| rat anti–α-tubulin antibody,tyrosinated, clone YL1/2 | Millipore Sigma | MAB1864-I | Antibody |
| Sodium Acrylate | Sigma | 408220 | ExM Gel |
| Streptavidin / Biotin blocking kit | Vector Laboratories | SP-2002 | Blocking buffer |
| Tris-HCl | Life Technologies | AM9855 | Digestion buffer |
| U2OS | ATCC | HTB-96 | Cell line |
| 6 well glass bottom plates | Cellvis | P06-1.5H-N | Imaging plate |
References
- Shi, X., et al. Label-retention expansion microscopy. The Journal of Cell Biology. 220 (9), 202105067 (2021).
- Chen, F., Tillberg, P. W., Boyden, E. S. Expansion microscopy. Science. 347 (6221), 543-548 (2015).
- Tillberg, P. W., et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nature Biotechnology. 34 (9), 987-992 (2016).
- Truckenbrodt, S., Sommer, C., Rizzoli, S. O., Danzl, J. G. A practical guide to optimization in X10 expansion microscopy. Nature Protocols. 14 (3), 832-863 (2019).
- Wang, Y., et al. Combined expansion microscopy with structured illumination microscopy for analyzing protein complexes. Nature Protocols. 13 (8), 1869-1895 (2018).
- Chozinski, T. J., et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nature Methods. 13 (6), 485-488 (2016).
- Ku, T., et al. Multiplexed and scalable super resolution imaging of three-dimensional protein localization in size adjustable tissues. Nature Biotechnology. 34 (9), 973-981 (2016).
- Gautier, A., et al. An engineered protein tag for multiprotein labeling in living cells. Chemistry & Biology. 15 (2), 128-136 (2008).
- Keppler, A., Pick, H., Arrivoli, C., Vogel, H., Johnsson, K. Labeling of fusion proteins with synthetic fluorophores in live cells. Proceedings of the National Academy of Sciences of the United States of America. 101 (27), 9955-9959 (2004).
- Thevathasan, J. V., et al. Nuclear pores as versatile reference standards for quantitative super resolution microscopy. Nature Methods. 16 (10), 1045-1053 (2019).
- Truckenbrodt, S., et al. X10 expansion microscopy enables 25-nm resolution on conventional microscopes. EMBO Reports. 19 (19), 45836 (2018).
- Damstra, H. G. J., et al. Visualizing cellular and tissue ultrastructure using Ten-fold Robust Expansion Microscopy (TREx). eLife. 11, 73775 (2021).
- M’Saad, O., Bewersdorf, J. Light microscopy of proteins in their ultrastructural context. Nature Communications. 11 (1), 3850 (2020).

