Visualizing Early Infection Sites of Rice Blast Disease (Magnaporthe oryzae) on Barley (Hordeum vulgare) Using a Basic Microscope and a Smartphone
Summary
This is a straightforward protocol of a barley leaf sheath assay using minimal reagents and common laboratory equipment (including a basic smartphone). The purpose is to visualize the early infection process of blast disease in labs without access to advanced microscopy equipment.
Abstract
Understanding how plants and pathogens interact, and whether that interaction culminates in defense or disease, is required to develop stronger and more sustainable strategies for plant health. Advances in methods that more effectively image plant-pathogen samples during infection and colonization have yielded tools such as the rice leaf sheath assay, which has been useful in monitoring infection and early colonization events between rice and the fungal pathogen, Magnaporthe oryzae. This hemi-biotrophic pathogen causes severe disease loss in rice and related monocots, including millet, rye, barley, and more recently, wheat. The leaf sheath assay, when performed correctly, yields an optically clear plant section, several layers thick, which allows researchers to perform live-cell imaging during pathogen attack or generate fixed samples stained for specific features. Detailed cellular investigations into the barley-M. oryzae interaction have lagged behind those of the rice host, in spite of the growing importance of this grain as a food source for animals and humans and as fermented beverages. Reported here is the development of a barley leaf sheath assay for intricate studies of M. oryzae interactions during the first 48 h post-inoculation. The leaf sheath assay, regardless of which species is being studied, is delicate; provided is a protocol that covers everything, from barley growth conditions and obtaining a leaf sheath, to inoculation, incubation, and imaging of the pathogen on plant leaves. This protocol can be optimized for high-throughput screening using something as simple as a smartphone for imaging purposes.
Introduction
Magnaporthe oryzae, the rice blast fungus, infects an assortment of grain crops, including barley, wheat, and rice1. This pathogen causes devastating diseases and poses a worldwide threat to these valuable crops, causing complete crop loss if not controlled. Many labs around the world focus on rice blast disease because of its global threat and its attributes as an excellent model for plant-fungal interactions2. It has been fully sequenced, and the genetics of its infective cycle, particularly the early events, have been established3,4. The life cycle begins with a spore germinating on a leaf surface, forming the specialized penetration structure called the appressorium. The appressorium penetrates the leaf tissue, and infection continues with the development of lesions which start the process of sporulation and spread disease4. Preventing any of these early events would drastically inhibit this devastating disease. Consequently, most current research on blast disease has been focused on the early infection steps, from the germinated conidia forming an appressorium to the development of the invasive hyphae and the biotrophic interfacial complex (BIC)5.
The vast amount of research on blast disease has been conducted in rice, even though M. oryzae is a significant pathogen for a variety of crops, and newly evolved strains are emerging as a global threat to wheat6. While rice is one of the top three staple crops used to feed the population, along with wheat and corn, barley is the fourth cereal grain in terms of livestock feed and beer production7. As the craft beer industry grows, so does the economic value of barley. There are distinct advantages of using M. oryzae and barley as a pathosystem to study blast disease. First, there are strains of M. oryzae that infect only barley, as well as strains that can infect multiple grass species. For example, 4091-5-8 infects primarily only barley, while Guy11 and 70-15 can infect both barley and rice8. These strains are genetically similar, and the infection process is comparable9. Second, under standard laboratory and greenhouse conditions, barley is easier to grow, as it doesn't have the complicated requirements of rice (concise temperature control, high humidity, specific light spectra). There are also imaging challenges with rice due to the hydrophobicity of the leaf surface, which barley does not exhibit10.
This protocol presents a simple method for isolating and effectively utilizing barley leaf sheaths for microscopic analysis of multiple infection stages, using common laboratory supplies and a smartphone for data collection. This method for the barley leaf sheath assay is adaptable for labs across the world as it requires minimal supplies, and yet provides a clear picture of the microscopic interaction between the pathogen and the first few cells it infects. Whereas pathogenicity assays, such as a spray or droplet inoculation, can provide a macro view of the pathogen's ability to form lesions, this assay allows the researcher to visualize specific steps of early infection, from pre-penetration events to colonization of epidermal cells. Further, researchers can easily compare infection with the wild-type fungus to infection with a mutant reduced in virulence.
Protocol
1. Preparation of experimental materials
- Prepare oatmeal agar (OMA) by blending oatmeal until it is a fine powder. Add 25 g of oatmeal powder and 15 g of agar to 500 mL of ddH2O, and autoclave on media cycle (alternatively bring to a boil for 20 min). Pour the media into sterile 60 mm Petri dishes.
NOTE: Other media types that induce sporulation, such as V8 agar, are acceptable for this protocol. - Plate M. oryzae filter stocks directly onto the OMA plates using sterile forceps, and allow them to cover the entire plate (9-12 days). Place the plates in a growth incubator at 25 °C with 12:12 h day:night cycles to help induce sporulation.
NOTE: Some mutants grow more slowly and require additional care (e.g., complete media first, then a transfer to the OMA), and could take an additional week to produce enough conidia. - Directly plant barley (Hordeum vulgare Lacey) seeds in a moist growth medium (e.g., soilless potting media) with 10-15 seeds per 6 inch pot. Place the pots in trays with 1-2 in of water.
- Set the barley growth chamber conditions as 22 °C for 12 h (daylight) and 19 °C for 12 h (dark) at 60% relative humidity. Continue to water from the bottom so as not to disturb the seeds.
- Grow the barley until the second leaf stage, approximately for 14 days. Using sterile scissors, cut the barley plant just above the soil line. Using forceps and a razor/scalpel, carefully cut the leaf sheath of the first leaf that is open longitudinally, and using the forceps, remove it from the base of the second leaf.
NOTE: Clean the tools (forceps, scalpel, etc.) before use with 75%-80% ethanol. The sheath is the thin epidermis layer that provides the attachment from the first to the second leaf (second to third leaf, etc.; see Figure 1). - Place the first leaf flat in a sterile 60 mm Petri dish, containing a wet paper towel to maintain humidity inside the plate. Using the scalpel, cut the majority of the first leaf away from the sheath, leaving only 0.5 in of the leaf tissue for mounting.
- Tape the leaf tissue to the bottom of the Petri plate.
NOTE: The sheath curls, but this is acceptable as a curled sheath holds the conidial droplet more easily. - Collect 9-12-day-old M. oryzae plates, and add 0.5-2 mL of sterile water to the plates. Using a sterile inoculation loop, gently scrape the mycelia to release the attached conidia. Carefully pipette the conidial suspension into a microcentrifuge tube containing a small piece of cheesecloth to filter out any large pieces of mycelium from the conidial suspension.
NOTE: Spores can be collected as early as 7 days, if growth and sporulation are sufficient to reach the desired spore concentration. Collection can be delayed no more than 14 days if working with a slow-growing genetic mutant - The desired spore concentration is 5 x 104 spores per mL, but a range (1 x 104-1 x 105) is acceptable. Too high of a concentration makes imaging individual infection sites challenging; dilute the spore concentration with sterile water if necessary.
- Carefully pipette the conidial suspension inside of the rolled leaf sheath. Start with 25 µL (the droplet size can be increased depending on the size of the sheath, up to 50 µL).
NOTE: It is recommended to perform three to five replicates of each mutant strain or barley line. It is common for damage to occur during the staining process, therefore additional sheath replicates are recommended. - Fill four or five 500 mL beakers with ddH2O, and heat until steaming (using a microwave or hotplate). Use caution when moving the hot water beakers. Hold the lid of the Petri dish over one of the steaming beakers to trap humidity inside the plate.
- Stack the infected leaf sheath plates and surround them with the remaining hot beakers. This creates a humid, moist environment, required for the spores to germinate.
NOTE: Use caution when steaming the lids to ensure no hot water or steam touches the sheaths. - Protect the leaf sheaths from light, cover with a solid colored (black preferred) rubber or plastic box, and let sit for 48 h or the desired time-point for imaging.
NOTE: A cardboard box is not suitable because it does not lock in the moisture/humidity and absorbs the steam from the hot water beakers. A locking, plastic tub works well for containing the humidity, and it can be covered with black fabric or a larger dark container to block the light.
2. Staining process
- Prepare the stain as follows: prepare a fresh dilution of 45% acetic acid and add 0.1% v/v trypan blue. Aliquot 1 mL of the dye solution into microcentrifuge tubes. Set a heat block or water bath to 40 °C.
- Carefully, using a razor or scalpel, cut the leaf sheath away from the tape. Using forceps, place the sheath into the microcentrifuge tube and ensure that it is completely submerged in the dye solution. Allow 2 h for the dye to penetrate the leaf. Heat the samples at 40 °C during the staining process time in a heat block or water bath to increase the penetration of the dye.
NOTE: The sheaths try to float in the micro-centrifuge tube; to prevent unstained pockets of leaf tissue, fill the tubes, submerge the sheaths, and close the tube. Do not put multiple sheaths in the same micro-centrifuge tube. - Rinse the leaf sheaths carefully in 60% glycerol to remove the extra dye. Three rinses (each in fresh glycerol) are generally sufficient. Keep the sheath in glycerol until ready to mount on slides.
3. Mounting and imaging process
- Place the sheath on a clean glass slide and add a few drops of 60% glycerol. Using a dissecting microscope and two pairs of forceps, carefully unroll the sheath, leaving the inoculated center facing up. Hold the sheath open with the forceps, and place the coverslip on top to prevent the sheath from curling and blocking the infection site.
NOTE: The leaf sheath is very fragile, and these steps need to be done with care to prevent damage to the sheath. - Seal the coverslip using nail polish for long-term storage, or tape for short-term storage. Observe the slides under a compound light microscope.
- Take basic images using a microscope and a cell phone. Here, images were taken with a cell phone adapter mount and a smartphone. For Android devices, adjust the camera application to the following settings: flash off, disable Top Shot,disable automatic adjustment of brightness and shadows, and set the photo resolution to full.
NOTE: Using the camera application on the phone decreases the battery life faster than usual, so having external power is recommended. - Once the cell phone is mounted on the microscope, take an image of a scale micrometer with the objective that will be used to acquire the data. The data in this study was acquired at a 40x 0.65 NA air objective, and the phone adapter was mounted on a 10x ocular. Adjust the zoom of the phone to 2.5x, and keep it consistent to maintain a constant pixel size.
- The center of the sheath houses the largest concentration of spores and infecting appressoria; therefore, aim for 9-12 images of each sheath to obtain significant numbers for statistical analysis. The number of spores and appressoria vary based on the concentration of spores applied.
4. Image assessment and counting using ImageJ (FIJI)
- Transfer the images to a computer running ImageJ (FIJI). To open the images, drag and drop the files onto the ImageJ bar.
- Set the scale of the images by loading the stage micrometer image and drawing a straight line between two markings for the scale. Open Set Scale, type in the Known Distance for the line measured, and type in the unit for the scale. On the micrometer, in this example, the smallest line was 10 µm. Check the Set Global box and hit OK. All subsequent images loaded will have the same scale.
- To count appressoria, spores, or other objects, select the Point tool. Next, open the ROI Manager. Click the T key on the keyboard to add points to the list. These regions of interest can be saved if needed and reloaded onto the same image.
- Depending on the experimental goals, make additional measurements, such as spore length, appressoria size, and germ tube length.
Representative Results
A depiction of the initial workflow for this technique is displayed in Figure 1. The sheaths were harvested from 14-day-old susceptible "Lacey" barley plants (H. vulgare). The conidia were harvested from 10-day-old sporulating M. oryzae OMA plates, with a conidial suspension prepared using sterile ddH2O for a final concentration of 5 x 104 spores per mL. The inoculum suspension was directly applied to the leaf sheaths, which were secured to sterile Petri plates. The plates were kept in a warm, humid chamber for 48 h without light. Following the incubation period, the leaf sheaths were stained with trypan blue and prepared for imaging.
Infection sites were imaged using a smartphone and smartphone microscope adapter. A minimum of 10 images were recorded for each of the tested strains of M. oryzae. The experiment was repeated three times for a minimum of 30 images for each fungal strain. Figure 2A shows the representative results of a successful sheath assay, along with unstained and improperly dyed images for reference.
Depending on the hypothesis, these images can be quantified and analyzed in a plethora of ways. For this experiment, at 48 h post-inoculation, the total number of live spores (germinated spores) were counted, along with the number of appressoria and the number of successfully infected cells. A collection of 2,000 randomly mutagenized M. oryzae strains in a barley-infecting background were generated in the lab. Pathogenicity assays using spray and drop inoculations revealed many mutants with reduced lesion size compared to the wild type (a common phenotype for M. oryzae mutants)11. To tease apart these phenotypes, it was hypothesized that the reduced lesion size was caused by inhibition of one of the early infection steps (spore germination, appressorial formation, penetration peg formation, initial epidermal cell colonization), which is most easily tested via the leaf sheath assay. A promising candidate from the mutagenesis project was identified using a forward genetic named J99A12. This mutant did not show sensitivity to either nitrogen-starved or reactive oxygen conditions during the screen. During follow-up experiments, J99A produced significant numbers of appressoria on a hydrophobic surface, but displayed reduced lesion size on live barley. When tested using the sheath assay, J99A successfully developed appressoria and penetration pegs, that penetrated the leaf sheath but did not produce invasive hyphae once inside, thus suggesting the infection stopped at the penetration peg (Figure 2B). Successfully infected cells were identified by the presence of infective hyphae inside the tissue of the leaf sheath. Comparing the number of appressoria to the number of infected cells provided a percentage of successfully infecting appressoria. For wild-type 4091-5-8, 87% of appressoria successfully invaded and colonized the cell, while in the mutant J99A, only 36% of appressoria had hyphae inside the cell12.
Figure 1: Leaf sheaths harvested from 10-day-old barley and carefully removed with tools cleaned in ethanol. Conidia are collected, and the concentration is adjusted to 0.5-1.0 x 105 per milliliter with sterile water. The isolated sheaths are taped inside of a 60 cm Petri plate, and the conidial suspension is loaded into the sheath. The inoculated samples are kept at room temperature, in the dark, with beakers of hot water for humidity. The samples are stained in 45% acetic acid + 0.1% trypan blue for 1-2 h at 40 °C, then rinsed in 60% glycerol three times for 48 h post-inoculation. Stained samples are mounted and imaged. Please click here to view a larger version of this figure.
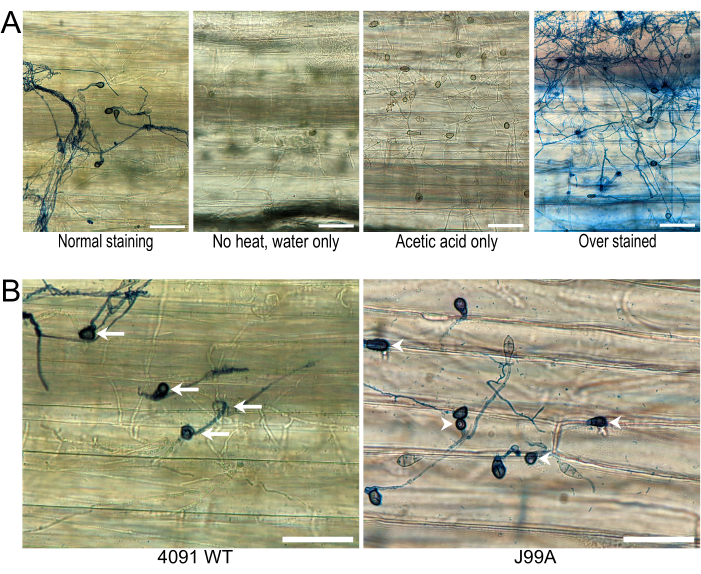
Figure 2: Representative results from the staining protocol. (A) Deviations from the staining protocol can result in suboptimal results. The heat and acetic acid serve to gently soften the leaf tissue. The post-staining rinses in 60% glycerol not only remove the excess stain, but help reduce the light scattering caused by the leaf and improve the image quality. Scale bars = 50 µm. (B) Representative images showing the robustness in this assay to see failed penetration and subsequent infection attempts of J99A (arrowheads), compared to successful attempts of 4091WT that resulted in the production of invasive hyphae to the leaf tissue (arrows). Scale bars = 50 µm. All images were taken 48 h post-infection. Please click here to view a larger version of this figure.
Discussion
There are many commonly used assays available to test M. oryzae strains that provide a macroscopic-level visual of a compatible or incompatible infection response, such as spray or droplet inoculations, and the use of rating systems to quantifylesion sizes13,14. Another common assay for M. oryzae is to test the ability of the pathogen to form its specialized penetration structure, the apppressorium15. Described here is an easy method for observing changes in early infection processes quickly and efficiently at the cellular level, in the more facile barley plant. This method is unique, in that it uses general lab equipment and a smartphone for data capture. This method negates the need for camera-equipped and computer-controlled microscopes, making this protocol affordable for any lab. Using this method, we were able to identify at which step mutant J99A infection was halted, a question previous experiments have been unable to clarify.
The infection process of M. oryzae in rice, particularly colonization of the first few epidermal cells, has been well-imaged using fluorescent proteins, markers, dyes, confocal imaging, and advanced microscopy16. These types of imaging experiments are expensive, time-consuming, and require specific expertise. Many labs, using homologous recombination, are able to create genetic mutants of M. oryzae to analyze individual gene’s roles in the infection cycle, but may not have access to the advanced equipment and expertise required to explore the underlying cell biology. This protocol is intended to help bridge this gap, by using only a compound light microscope and a smartphone to capture digital images and generate z-stack-type videos of fixed leaf sheath tissues. This method enables imaging a few cell layers into the tissue, capturing the invasive fungal hyphae for up to 48 h. The leaf sheath of rice and barley have similar characteristics; they are only a few cell layers thick and have less chloroplasts, making them easier to image. As stated above, barley is less hydrophobic and easier to grow than rice, and many strains of M. oryzae can cause infection on rice and barley, making this experiment an easy swap for the more complicated rice assays.
A few limitations of this method include using the video function of the smartphone to collect z-field data, as the z-increment for the frame rate is unknown. Another limitation is that it requires fixed tissue (not live cells). However, because of the speed and ease of the protocol, this limitation could be overcome by examining various time points post-infection.
One of the most crucial steps of the protocol is proper staining. Improper rinsing causes excess dye in the slide preparation, causing dye saturation, and making the pathogen tissue indistinguishable from the leaf tissue. Meanwhile, under-staining prevents the pathogen tissue from contrasting against the leaf tissue.
The aforementioned method is adaptable to many scientific questions, and can be used to assess fungal mutants, assess various degrees of genetics resistance in barley plants, and test the efficiency of previously applied fungal control methods. It is also possible to extend this method to other plant-pathogen interactions, particularly other monocot leaf sheaths.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors acknowledge funding from the USDA-NIFA award 2016-67013-24816.
Materials
| Acetic acid | Sigma-Aldrich | A6283 | |
| Cell phone | Pixel 4A | Any smartphone with a rear facing camera that can be mounted in an a holder will suffice. | |
| Cell phone Microscope adapter | Vankey | B01788LT3S | https://www.amazon.com/Vankey-Cellphone-Telescope-Binocular-Microscope/dp/B01788LT3S/ref=sr_1_2_sspa?keywords=vankey+cellphone+telescope+adapter+mount&qid=1662568182&sprefix= vankey+%2Caps%2C63&sr=8-2 -spons&psc=1&spLa=ZW5jcnlwd GVkUXVhbGlmaWVyPUFKNklBR jlCREJaMEcmZW5jcnlwdGVkSWQ 9QTA2MDMxNjhBRFYxQTMzNk9E M0YmZW5jcnlwdGVkQWRJZD1BM DQxMzAzOTMxNzI1TzE3M1ZGTEI md2lkZ2V0TmFtZT1zcF9hdGYmY WN0aW9uPWNsaWNrUmVkaXJlY3 QmZG9Ob3RMb2dDbGljaz10cnVl |
| Glycerol | Sigma-Aldrich | G5516 | |
| Microscope | AmScope | FM690TC | 40x–2500x Trinocular upright epi-fluorescence microscope |
| Oatmeal old fashioned rolled oats | Quaker | N/A | https://www.amazon.com/Quaker-Oats-Old-Fashioned-Pack/dp/B00IIVBNK4/ref=asc_df_B00IIVBNK4/?tag=hyprod-20&linkCode=df0 &hvadid=312253390021&hvpos= &hvnetw=g&hvrand=98212627704 6839544&hvpone=&hvptwo=&hvq mt=&hvdev=c&hvdvcmdl=&hvlocint =&hvlocphy=9007494&hvtargid =pla-568492637928&psc=1 |
| ProMix BX | ProMix | 1038500RG | |
| Rectangular coverglass | Corning | CLS2975245 | |
| Slides, microscope | Sigma-Aldrich | S8902 | |
| Stage micrometer | OMAX | A36CALM7 | 0.1 mm and 0.01 mm Microscope calibration slide |
| Trypan blue | Sigma-Aldrich | T6146 |
References
- Roy, K. K., et al. First report of barley blast caused by Magnaporthe oryzae pathotype Triticum (MoT) in Bangladesh. Journal of General Plant Pathology. 87 (3), 184-191 (2021).
- Dean, R., et al. The Top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology. 13 (4), 414-430 (2012).
- Dean, R. A., et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 434 (7036), 980-986 (2005).
- Wilson, R. A., Talbot, N. J. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nature Reviews. Microbiology. 7 (3), 185-195 (2009).
- Giraldo, M. C., et al. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nature Communications. 4, 1996 (2013).
- Islam, M. T. Emergence of wheat blast in Bangladesh was caused by a SouthAmerican lineage of Magnaporthe oryzae. BMC Biology. 14 (1), 84 (2016).
- Langridge, P. Economic and Academic Importance of Barley. The Barley Genome. Compendium of Plant Genomes. , 1-10 (2018).
- Heath, M. C., Valent, B., Howard, R. J., Chumley, F. G. Interactions of two strains of Magnaporthe grisea with rice, goosegrass, and weeping lovegrass. Canadian Journal of Botany. 68 (8), 1627-1637 (1990).
- Gowda, M., et al. Genome analysis of rice-blast fungus Magnaporthe oryzae field isolates from southern India. Genomics Data. 5, 284-291 (2015).
- Luginbuehl, L. H., El-Sharnouby, S., Wang, N., Hibberd, J. M. Fluorescent reporters for functional analysis in rice leaves. Plant Direct. 4 (2), 00188 (2020).
- Fernandez, J., Wilson, R. A. Why no feeding frenzy? Mechanisms of nutrient acquisition and utilization during infection by the rice blast fungus Magnaporthe oryzae. Molecular Plant-Microbe Interactions. 25 (10), 1286-1293 (2012).
- Cooper, J. G. Identifying Genetic Control of Reactive Oxygen Species in Magnaporthe oryzae (the Rice Blast Fungus) through Development, Screening, and Characterization of a Random Insert Mutant Library. University of Delaware. , (2022).
- Zhang, M., et al. al.The plant infection test: Spray and wound-mediated inoculation with the plant pathogen Magnaporthe grisea. Journal of Visualized Experiments. (138), e57675 (2018).
- Koga, H., Dohi, K., Nakayachi, O., Mori, M. A novel inoculation method of Magnaporthe grisea for cytological observation of the infection process using intact leaf sheaths of rice plants. Physiological and Molecular Plant Pathology. 64 (2), 67-72 (2004).
- Hamer, J. E., Howard, R. J., Chumley, F. G., Valent, B. A mechanism for surface attachment in spores of a plant pathogenic fungus. Science. 239 (4837), 288-290 (1988).
- Khang, C. H., et al. et al. of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. The Plant Cell. 22 (4), 1388-1403 (2010).

