Methods for Study of Neuronal Morphogenesis: Ex vivo RNAi Electroporation in Embryonic Murine Cerebral Cortex
Summary
To conduct a rapid assessment of the function of genes in the development of cerebral cortex, we describe methods involving the ex vivo electroporation of plasmids co-expressing inhibitory RNA (RNAi) and GFP in murine embryonic cortex. This protocol is amenable to the study of various aspects of neurodevelopment such as neurogenesis, neuronal migration and neuronal morphogenesis including dendrite and axon outgrowth.
Abstract
The cerebral cortex directs higher cognitive functions. This six layered structure is generated in an inside-first, outside-last manner, in which the first born neurons remain closer to the ventricle while the last born neurons migrate past the first born neurons towards the surface of the brain1. In addition to neuronal migration2, a key process for normal cortical function is the regulation of neuronal morphogenesis3. While neuronal morphogenesis can be studied in vitro in primary cultures, there is much to be learned from how these processes are regulated in tissue environments.
We describe techniques to analyze neuronal migration and/or morphogenesis in organotypic slices of the cerebral cortex4,6. A pSilencer modified vector is used which contains both a U6 promoter that drives the double stranded hairpin RNA and a separate expression cassette that encodes GFP protein driven by a CMV promoter7-9. Our approach allows for the rapid assessment of defects in neurite outgrowth upon specific knockdown of candidate genes and has been successfully used in a screen for regulators of neurite outgrowth8. Because only a subset of cells will express the RNAi constructs, the organotypic slices allow for a mosaic analysis of the potential phenotypes. Moreover, because this analysis is done in a near approximation of the in vivo environment, it provides a low cost and rapid alternative to the generation of transgenic or knockout animals for genes of unknown cortical function. Finally, in comparison with in vivo electroporation technology, the success of ex vivo electroporation experiments is not dependant upon proficient surgery skill development and can be performed with a shorter training time and skill.
Protocol
1. Preparing Culture Solutions and Media (not in video)
- Prepare 1 liter of Complete Hank’s balanced salt solution (HBSS) containing 1x HBSS, 2.5 mM Hepes (pH7.4), 30 mM D-glucose, 1 mM CaCl2, 1 mM MgSO4 and 4 mM NaHCO3. Add double distilled water (ddH2O). Filter sterilize with a 0.2-μm filter and store at 4 °C.
- Prepare slice culture medium using 35 mL of Basal Medium Eagle media, 12.9 mL of Complete HBSS, 20 mM D-glucose (1.35 mL of 1 M solution), 1 mM Glutamax (0.25 mL of a 200 mM solution), 0.5 ml of Penicillin-streptomycin 100x stock. Filter sterilize with a 0.2-μm filter, then add heat-inactivated horse serum to a final concentration of 5%.
- Prepare Laminin working solution by making a 1 mg/mL Laminin stock solution with sterile distilled deionized water. Prepare 100 μL aliquots in 0.5 mL eppendorf tubes and freeze at -80 °C.
- Prepare Poly-L-lysine working solution by adding 5 mL of sterile H2O to 5 mg of poly-L-lysine to make a 1 mg/mL stock solution. Prepare 1 mL aliquots and freeze at -20 °C.
- Prepare coating solution by diluting 1 mL of poly-L-lysine and 100 μL of laminin to a final volume of 12 mL with sterile water. Make this solution fresh each time.
2. Preparing Organotypic Slice Inserts (not in video)
- Prepare two six-well plates with one culture insert per well using sterile forceps. Add 2 mL of sterile ddH2O underneath the culture inserts.
- Add 1 mL of the coating solution on top of the membrane taking care not to puncture the membrane. Incubate overnight in a humidified incubator at 37 °C and 5% CO2.
- Remove coating media and wash membrane with sterile H20 three times. Let inserts dry before use. Add 1.8 mL of slice culture medium and place in 37 °C incubator. Wrap the unused plates with Parafilm and store at 4 °C for up to 4 weeks.
3. Preparing for Electroporation (not in video)
- Prepare RNAi constructs for electroporation. Double strand RNA hairpin inserts were cloned into a pSilencer vector. The plasmid contains: 1) a U6 promoter that drives the double strand RNA generation; and 2) a GFP expression cassette driven by the CMV promoter8,9. This plasmid has been previously described by Konishi and colleagues9 and others7,8. Plasmids are purified using a Qiagen maxi-prep kit, and used at a concentration of 1 mg/mL.
- In order to visualize DNA while injecting it, prepare a 0.5% fast green dye solution and use it at 1:20 with the DNA to be injected (usually 20 μL of DNA with 1 μL of fast green). Left over DNA-fast green dye mixture can be stored at -20 °C for up to a week.
- Clean dissection area, dissecting tools and vibratome vessel with 70% ethanol. Chill complete HBSS so that it is ice cold. Chill vibratome vessel by packing ice around it inside the vibratome with some water for rapid cooling. Prepare 3% low melting point agarose using complete HBSS. Microwave for 1 min. Avoid over boiling. Keep in a 42 °C waterbath until use.
- Electroporation parameters are set as follows. For an E15 embryo use 35 V, 5 pulses, 100 ms length, 900 ms interval between pulses. For older animals, to ensure electroporation, use higher voltage up to 50 V or increase the number of pulses up to 8 pulses. To prevent damaging the tissue in younger animals, use either fewer pulses or up to 2 pulses and lower voltage up to 25 V. These parameters may be varied and determined empirically depending on the age of the animal.
4. Dissection and Electroporation (in video)
- After euthanizing a pregnant female, dissect embryos out into ice cold complete HBSS. Keep each embryo in their individual placental sacs.
- Dissect embryo out and cut off the head after the first vertebra. Keep in ice cold complete HBSS.
- For injection, place the head on a piece of Parafilm on top of a petri dish. Using custom made Hamilton syringe (see Table I) inject about 6 to 8 μL of DNA:fast green dye mix through the third ventricle in order to fill in both lateral ventricles in the cortical vesicles. Alternatively, injection can be done directly into each lateral ventricle.
- For the ex vivo electroporation, use BTX-tweezer platinum electrodes. Place the positive electrode towards the side of the cortex you want to electroporate i.e. top of the head for dorsal cortex.
- After electroporation, incubate the heads on ice for at least 5 min before dissecting.
- Dissect brains in ice cold HBSSby making a small incision on the side of the head and peeling the skin off the sides of the head. Next, with fine forceps gently peel away the pia from the brain. Remove the intact brain from the skull, taking care not to damage the cortex.
5. Embedding and Sectioning of Electroporated Cortices (in video)
- Transfer 3% low melting point agarose into a large mold placed on ice. The bottom of the mold will start to solidify faster which will prevent brains from sinking down to the bottom of the mold. Gently, transfer brains with fine forceps one by one after removing excess buffer with a Kimwipe or filter paper. Use a 10 μL pipet tip to swirl the brains inside the mold to ensure maximum interface between the agarose and brain tissue.
- Orient the brains to ensure all brains are in the same orientation and at about the same level in the agarose. Let the agarose solidify for about 5 min. Use bonding adhesive (crazy glue) to attach the agarose blocks so that the olfactory bulbs are standing up. Once the blocks are attached, immediately add ice cold HBSS and trim the agarose to make sure individual slices are obtained for each brain.
- To slice the blocks, set the vibratome’s velocity to a low speed (about half the maximum) and set the blade vibration frequency at the highest setting. Generate 250-μm thick coronal slices. Retrieve slices using a bent fine spatula and transfer them to tissue wells with a fine brush or forceps.
- In tissue culture hood, transfer slices into coated inserts. Add 500 μL of slice culture media to each insert to make the transfer easy. Up to 5 slices can be placed per insert. Remove excess media from the top of the slices and incubate at 37 °C in humidified incubator.
6. Culture and Analysis of Organotypic Slices (in video)
- In order to maintain healthy slices, fresh media should be added at least every other day underneath the membrane by replacing half of the media each time.
- In order to analyze slices after the desired days in culture, fix slices in the membrane. Wash with 1x phosphate saline buffer (PBS) at 37 °C three times for 10 min each time. Next, fix with 4% Paraformaldehyde (PFA) overnight at 4 °C or for 1 hr at room temperature.
- Slices can be analyzed with different cellular markers or stained with Hoechst alone to visualize electroporated and non-electroporated cells. Permeabilize and block slices for 2 hr at room temperature with 10% goat serum, 0.1% triton in 1x PBS with gentle shaking.
- Stain with Hoechst for 1 hr at room temperature, wash 3 times with 1x PBS 10 min each time with gentle shaking.
- To mount slices cut the membrane with a scalpel, and use a fine forceps to transfer the membrane containing slices to a frosted glass slides in a water chamber. Up to 5 slices can be placed per glass slide. Remove excess water, and add a drop of Flourmount solution to each brain slice. Gently place a coverslip on top of the brain slices and remove any air bubbles. Analyze slices using a confocal microscope.
7. Alternative Paraffin Embedding of Organotypic Slices (not in video)
- Organotypic slices can also be embedded for paraffin for a finer morphological analysis, to this end the membrane containing the organotypic slices can be fixed in 4% PFA as described above.
- The fixed slices are embedded in 1% agarose (pre-warmed to 37 °C), and solidified in ice for 30 min. The agarose blocks can then be post-fixed in 4% PFA at 4 °C for 30 min.
- The agarose block containing the organotypic slice will then be embedded in paraffin and processed for immunofluorescence as previously described10.
8. Representative Results
A diagrammatic representation of the electroporation of murine cortex and culture of organotypic slices is shown in Figure 1. This method is a useful strategy for rapid assessment of the function of genes involved in neuronal development11. Depending on the amount of DNA electroporated and the embryonic stage at electroporation, the transfection efficiency will vary. Slices will start expressing GFP at least 8 hours post-electroporation and cells will undergo the normal sequence of neurogenic events (proliferation, migration, and early neuronal differentiation) in culture. Figure 2 shows an electroporated brain slice that is expressing a control pSilencer- GFP vector and one can observe neuronal progenitors, migrating neurons and differentiated neurons in the slice. Organotypic slices will keep their morphology as long as they are maintained in a good media-air interface on the membranes and can be used up to at least 5 days in culture.
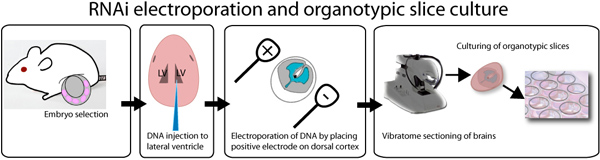
Figure 1. Illustration of ex vivo electroporation and organotypic slice culture assay. E14.5 embryos are dissected out, and individually injected with DNA mixed with fast green dye in order to visualize the injection site. DNA can be injected in both the lateral ventricles as depicted in the illustration or in the third ventricle in order to fill the lateral ventricles. After injection, brains are electroporated with a square wave electroporator, placing the positive electrode on the desired side of the brain. Brains are embedded in 3% low melting point agarose and sectioned using a vibratome. 250 μm brain slices are placed on 0.4 μm inserts and cultured up to a week. GFP can be observed after 8 hours post transfection.
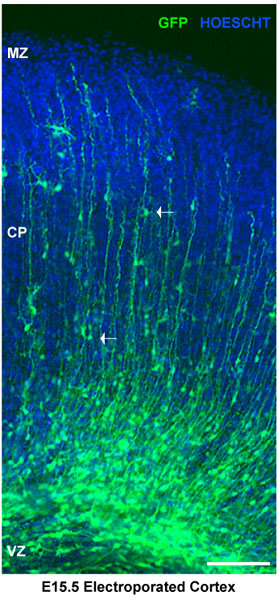
Figure 2. Analysis of electroporated brain slices. Electroporated brain slices were stained for Hoescht. Dorsal cortex shows electroporated neuronal progenitors at the ventricular zone (vz). Neurons in the cortical plate (cp) are delimited by the marginal zone (mz). White arrows show migrating neurons. In this case, brains were injected at E15.5 and electroporated with a pSilencer GFP control vector. The sections represent cortical explants 4 days after electroporation. Scale bar 100 μm.
Troubleshooting:
- Low transfection efficiency: Adjust the concentration of DNA used to at least 1 μg/μL. Always use very clean DNA from a maxi prep if necessary use an endo-free Quiagen kit to purify the DNA.
- Cells transfected in a different brain area than the one desired: Ensure that the electrodes are positioned correctly with the positive electrode position towards the side of the brain to be electroporated.
- Organotypic slices lose morphology: Change media every day and ensure the slices are not floating in media
- Slices come off the agarose as they are being cut in the vibratome: Ensure that a good interface is made when embedding the brains in the low melting point agarose.
Discussion
These methods involving the ex vivo electroporation of plasmids encoding double stranded RNA hairpins 8 and culture of organotypic slices4 provide several distinct advantages. First, these methods allow for a rapid assessment of RNAi-derived phenotypes. The inclusion of an expression cassette encoding GFP in the same pSilencer vector that contains the U6 promoter that drives the double stranded RNA hairpin allows for a rapid identification and characterization of cells electroporated with the RNAi vector. In addition to time-efficiency, these methods are highly cost-effective compared to the generation of several knockout lines. Second, in comparison with survival electroporation technology12 that requires longer training and skill development to ensure survival of the animal and success of the electroporation, methods described here can be performed with a shorter training time and skill. The in vivo survival surgeries are also mired by a baseline failure rate that may be associated with animal (mother and pup) morbidity. These animals will frequently need to be assessed by reasonably skilled staff and/or veterinarians from the animal facility. Third, because they are cultured for up to 5 days or more in vitro,they allow us to address a wide array of questions related to neuronal proliferation, cell fate determination, neuronal migration and the early stages of neuronal maturation (neurite outgrowth). Fourth, because we are working in culture, we are also able to add exogenous factors to test the role of growth factors and pharmaceutical agents in the neurodevelopmental mechanisms described. Fifth, the electroporation approach ex-vivo is preferable to a gene gun approach because it allows targeting of specific regions of the brains by precisely orienting the electrodes. Finally, compared to viral transduction of organotypic slices the electroporation approach allows for a higher transfection efficiency of primary cells.
These methods do have a few limitations. Unless the slice technique is modified to sustain longer culture periods, these methods may not be ideal to assess synaptogenesis or developmental events that occur later. An additional limitation may be that the reproducibility of the results is highly dependent on electrode placement. Some practice is required to ensure electroporation of the same brain region which increases reproducibility of results. Finally, the maintenance of the air/liquid interface is also crucial to ensure healthy slices. As described above, we anticipate that these skills can be easily acquired by the JoVE readers, and that the strengths of these methods greatly outweigh the limitations given the appropriate application. In summary, vertebrate neurodevelopment is controlled by a large number of genes. Neuronal morphogenesis in particular is a very interesting and important topic. This approach allows for a highly efficient method for screening gene function during neurodevelopment and may serve a very broad range of experimental purposes.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Dr. Shirin Bonni for providing the pSil-GFP construct, Dr. Alper Uzun for the illustration of Figure 1, and the Leduc Bioimaging facility for confocal microscopy. EMM is supported by the Career Award for Medical Science from the Burroughs Wellcome Fund, a NARSAD Young Investigators Award, and NIH NCRR COBRE P20 RR018728-01. SBL is supported by NIH NCRR COBRE P20 RR018728-01, and has received support from PHS NRSA 5T32MH019118-20.
Materials
| Name of the reagent | Company | Catalogue number | Comments |
| Hamilton syringe | HAMILTON | 80008 | 31 gauge, 0.5 inches long,PT-4 (level of point beveling), 10μl volume |
| Platinum tweezertrodes | BTX | 45-0489 | 5mm size |
| ECM830 electroporator | BTX | 45-0002 | |
| BTX Footswitch | BTX | 45-0208 | For use with ECM830 electroporator |
| Vibrating blade microtome | LEICA | VT1000 S | |
| 6- well dish to use with inserts | FALCON | 353502 | Contains notches to fit inserts |
| Tissue culture inserts | FALCON | 353090 | 0.4 micrometer |
| Fast Green | SIGMA | F7252 | |
| Low Melting Agarose | FISHER | BP165-25 | DNA grade |
| Laminin | Sigma-Aldrich | L2020 | |
| Poly-L-lysine | Sigma-Aldrich | P5899 | |
| Basal Medium Eagle | Sigma Aldrich | B-1522 | |
| HBSS 10x without Ca and Mg | GIBCO | 14180-046 | |
| HEPES-free acid | Sigma- Aldrich | H4034 |
References
- Angevine, J. B., Sidman, R. L. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 192, 766-766 (1961).
- Kriegstein, A. R., Noctor, S. C. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 27, 392-392 (2004).
- Barnes, A. P., Polleux, F. Establishment of axon-dendrite polarity in developing neurons. Annu. Rev. Neurosci. 32, 347-347 (2009).
- Haydar, T. F., Bambrick, L. L., Krueger, B. K., Rakic, P. Organotypic slice cultures for analysis of proliferation, cell death, and migration in the embryonic neocortex. Brain Res. Brain Res. Protoc. 4, 425-425 (1999).
- Polleux, F., Ghosh, A. The slice overlay assay: a versatile tool to study the influence of extracellular signals on neuronal. Sci. STKE. 2002, pl9-pl9 (2002).
- Guerrier, S. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 138, 990-990 (2009).
- Stegmuller, J. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron. 50, 389-389 (2006).
- Sepp, K. J. Identification of neural outgrowth genes using genome-wide RNAi. PLoS Genet. 4, e1000111-e1000111 (2008).
- Konishi, Y. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 303, 1026-1026 (2004).
- Vankelecom, H. Fixation and paraffin-embedding of mouse tissues for GFP visualization. Cold Spring Harb Protoc. 2009, 5298-5298 (2009).
- Hand, R. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 48, 45-45 (2005).
- Taniguchi, Y., Young-Pearse, T., Sawa, A., Kamiya, A. In Utero Electroporation as a Tool for Genetic Manipulation in Vivo to Study Psychiatric Disorders: From Genes to Circuits and Behaviors. Neuroscientist. 18, 169-179 (2012).

