利用 Crispr-ca 在哺乳动物细胞系中进行基因组编辑
Summary
Crispr-卡斯是一种强大的技术, 用于设计复杂的植物和动物基因组。在这里, 我们详细介绍了一个使用不同的 Cas 内切酶有效编辑人类基因组的协议。我们强调重要的注意事项和设计参数, 以优化编辑效率。
Abstract
聚集定期间隔短回文重复 (CRISPR) 系统在细菌适应性免疫中自然发挥作用, 但已成功地将其重新用于许多不同生物的基因组工程。最常见的是, 与通配的通配型 CRISPR (Cas9) 或 Cas12a 内切酶用于裂解基因组中的特定位点, 之后 DNA 双链断裂通过非同源端连接 (NHEJ) 途径或同源定向修复 (HDR) 路径取决于捐献者模板是否分别不存在或存在。到目前为止, 来自不同细菌物种的 CRISPR 系统已被证明能够在哺乳动物细胞中进行基因组编辑。然而, 尽管该技术表面上很简单, 但仍需要考虑多个设计参数, 这往往会让用户对如何最好地进行基因组编辑实验感到困惑。在这里, 我们描述了一个完整的工作流程, 从实验设计到检测携带所需 DNA 修饰的细胞克隆, 目的是促进在哺乳动物细胞系中成功执行基因组编辑实验。我们强调了用户需要注意的关键注意事项, 包括 CRISPR 系统的选择、间隔长度以及单链寡核苷酸 (ssODN) 供体模板的设计。我们设想, 这种工作流程将有助于基因敲除研究、疾病建模工作或记者细胞系的生成。
Introduction
设计任何生物基因组的能力都有许多生物医学和生物技术应用, 例如纠正致病突变、构建准确的疾病研究细胞模型或农业发电具有理想性状的作物。自世纪之交以来, 哺乳动物细胞的基因组工程开发了各种技术, 包括美加努丁 1、2、3、锌指核酸酶4、5或转录激活物样效应酶 (talens)6,7,8,9。然而, 这些早期的技术要么难以编程, 要么繁琐组装, 从而阻碍了它们在研究和行业中的广泛采用。
近年来, 聚集定期间隔的短回文重复 (CRISPR)-crispr 相关 (Cas) 系统已成为一种强大的新的基因组工程技术 10,11。它最初是细菌中的适应性免疫系统, 已成功地用于植物和动物 (包括人类) 的基因组修饰。Crispr-ca 在如此短的时间内获得如此广泛的欢迎的一个主要原因是, 将关键的 Cas 内切酶 (如 Cas9 或 Cas12a) 带到基因组中正确位置的元素只是一个简短的嵌套单导 RNA (sgRNA), 设计简单, 合成便宜。在被招募到目标位点后, 卡斯酶起到了分子剪刀的作用, 并将其与鲁夫 c、hnh 或 nok 域 12、13、14 的结合 dna 裂解。由此产生的双链断裂 (DSB) 随后由细胞通过非同源端部连接 (NHEJ) 或同源定向修复 (HDR) 途径进行修复。在没有修复模板的情况下, DSB 通过容易出错的 NHEJ 途径进行修复, 这可能会导致在切割部位伪随机插入或删除核苷酸 (indels), 从而有可能导致蛋白质编码基因的帧-裂隙突变。然而, 在存在包含所需 DNA 变化的捐献者模板的情况下, DSB 通过高保真 HDR 途径进行修复。常见的供体模板类型包括单链寡核苷酸 (Ssodn) 和质粒。前者通常用于预期的 DNA 变化较小 (例如, 单个碱基对的变化), 而后者通常用于如果你希望插入一个相对较长的序列 (例如, 绿色荧光蛋白的编码序列或GFP) 进入目标位点。
卡斯蛋白的内切酶活性要求目标地点15 上存在一个基旋的相邻母题 (pam)。Cas9 的 PAM 位于原相的 3 ‘ 末端, 而 Cas12a 的 PAM (也称为 Cpf1) 位于 5 ‘ 端, 而不是16 ‘.如果 pam 不存在 17, 则 Cas-gue RNA 复合体无法引入 DSB.因此, PAM 对特定的 Cas 核酸酶能够裂解的基因组位置施加了约束。幸运的是, 来自不同细菌种类的 Cas 核酸酶通常表现出不同的 PAM 要求。因此, 通过将各种 CRISPR-Cas 系统集成到我们的工程工具箱中, 我们可以扩大可能在基因组中针对的站点的范围。此外, 可以设计或进化天然 cas 酶来识别替代 pam 序列, 从而进一步扩大了可用于操作18、19、20的基因组目标的范围。
虽然有多个 Crispr-ca 系统可用于基因组工程目的, 但由于多种原因, 该技术的大多数用户主要依赖来自化脓性链球菌(spcas9) 的 cas9 珠。首先, 它需要一个相对简单的 NGG PAM, 不像许多其他的钙蛋白, 只能在更复杂的 Pam 存在的情况下裂解。其次, 这是第一个成功部署在人的牢房21、22、23、24 的卡斯内切酶。第三, SpCas9 是迄今为止最有特色的酶。如果研究人员希望使用另一个 Cas 核酸酶, 他或她往往不清楚如何最好地设计实验, 以及与 SpCas9 相比, 其他酶在不同的生物环境中的表现如何。
为了清楚地了解不同 Crispr-ca 系统的相对性能, 我们最近对五个 Cas 内切酶–SpCas9、来自金黄色葡萄球菌的 cas9 酶 (sacas9)、cas9 酶进行了系统的比较。脑膜炎奈瑟菌 ( nmcas9), 来自 BV3L6 (ascas12a) 的 cas12a 酶, 以及来自Lachnospiraceae Nd2006 (LbCas12a 25 的 cas12a 酶. 为了进行公平的比较, 我们使用同一组靶点和其他实验条件对各种 Ca 核酸酶进行了评估。这项研究还描述了每个 Crispr-卡斯系统的设计参数, 这将为该技术的用户提供有用的参考。在这里, 为了使研究人员能够更好地利用 Crispr-卡斯系统, 我们提供了一个逐步协议, 使用不同的 Cas9 和 Cas12a 酶优化基因组工程 (参见图 1)。该协议不仅包括实验细节, 而且还包括重要的设计考虑, 以最大限度地提高在哺乳动物细胞中成功的基因组工程结果的可能性。
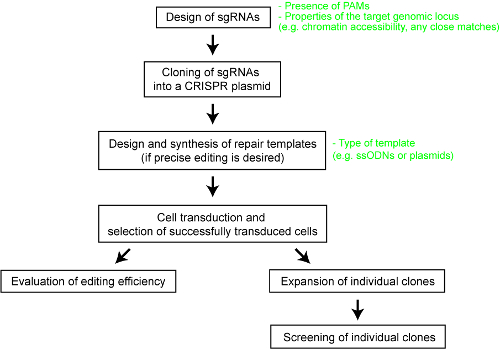
图 1: 生成经过基因组编辑的人类细胞系的工作流程概述.请点击这里查看此图的较大版本.
Protocol
Representative Results
Discussion
Crispr-卡斯系统是一种强大的革命性技术, 用于设计植物和动物的基因组和转录体。许多细菌物种已被发现含有 Crispr-卡斯系统, 这可能被用于基因组和转录组工程目的44。虽然来自化脓性链球菌(spcas9) 的 cas9 内切酶是第一个成功地在人体细胞 21,22,23,24的酶从其他细菌中应用物种,…
Disclosures
The authors have nothing to disclose.
Acknowledgements
M. h. t. 得到科学技术和研究机构联合理事会办公室赠款 (1431AFG103)、国家医学研究委员会赠款 (ofirg额 0017 2016)、国家研究基金会赠款 (NRF2013-THE001-0046 和 NRF2013 THEE001-001-00-93) a a 的支持。教育部一级赠款 (rgfa17 (s)), 来自南洋理工大学的创业补助金, 以及南洋理工大学为国际基因工程机 (iGEM) 竞赛提供的资金。
Materials
| T4 Polynucleotide Kinase (PNK) | NEB | M0201 | |
| Shrimp Alkaline Phosphatase (rSAP) | NEB | M0371 | |
| Tris-Acetate-EDTA (TAE) Buffer, 50X | 1st Base | BUF-3000-50X4L | Dilute to 1X before use. The 1X solution contains 40 mM Tris, 20 mM acetic acid, and 1 mM EDTA. |
| Tris-EDTA (TE) Buffer, 10X | 1st Base | BUF-3020-10X4L | Dilute to 1X before use. The 1X solution contains 10 mM Tris (pH 8.0) and 1 mM EDTA. |
| BbsI | NEB | R0539 | |
| BsmBI | NEB | R0580 | |
| T4 DNA Ligase | NEB | M0202 | 400,000 units/ml |
| Quick Ligation Kit | NEB | M2200 | An alternative to T4 DNA Ligase. |
| Rapid DNA Ligation Kit | Thermo Scientific | K1423 | An alternative to T4 DNA Ligase. |
| Zero Blunt TOPO PCR Cloning Kit | Thermo Scientific | 451245 | The salt solution comes with the TOPO vector in the kit. |
| NEBuilder HiFi DNA Assembly Master Mix | NEB | E2621L | Kit for Gibson assembly. |
| One Shot Stbl3 Chemically Competent E.Coli | Thermo Scientific | C737303 | |
| LB Broth (Lennox), powder | Sigma Aldrich | L3022 | Reconstitute in ddH20, and autoclave before use |
| LB Broth with Agar (Lennox), powder | Sigma Aldrich | L2897 | Reconstitute in ddH20, and autoclave before use |
| SOC media | – | – | 2.5 mM KCl, 10 mM MgCl2, 20 mM glucose in 1 L of LB Broth |
| Ampicillin (Sodium), USP Grade | Gold Biotechnology | A-301 | |
| REDiant 2X PCR Mastermix | 1st Base | BIO-5185 | |
| Agarose | 1st Base | BIO-1000 | |
| T7 Endonuclease I | NEB | M0302 | |
| Plasmid DNA Extraction Miniprep Kit | Favorgen | FAPDE 300 | |
| Dulbecco's Modified Eagle Medium (DMEM), High Glucose | Hyclone | SH30081.01 | 4.5 g/L Glucose, no L-glutamine, HEPES and Sodium Pyruvate |
| L-Glutamine, 200mM | Gibco | 25030 | |
| Penicillin-Streptomycin, 10, 000U/mL | Gibco | 15140 | |
| 0.25% Trypsin-EDTA, 1X | Gibco | 25200 | |
| Fetal Bovine Serum | Hyclone | SV30160 | FBS is heat inactivated before use at 56 oC for 30 min |
| Phosphate Buffered Saline, 1X | Gibco | 20012 | |
| jetPRIME transfection reagent | Polyplus Transfection | 114-75 | |
| QuickExtract DNA Extraction Solution, 1.0 | Epicentre | LUCG-QE09050 | |
| ISOLATE II Genomic DNA Kit | Bioline | BIO-52067 | An alternative to QuickExtract |
| Q5 High-Fidelity DNA Polymerase | NEB | M0491 | |
| Deoxynucleotide (dNTP) Solution Mix | NEB | N0447 | |
| 6X DNA Loading Dye | Thermo Scientific | R0611 | 10 mM Tris-HCl (pH 7.6) 0.03% bromophenol blue, 0.03% xylene cyanol FF, 60% glycerol, 60 mM EDTA |
| Protease Inhibitor Cocktail, Set3 | Merck | 539134 | |
| Nitrocellulose membrane, 0.2µm | Bio-Rad | 1620112 | |
| Tris-glycine-SDS buffer, 10X | Bio-Rad | 1610772 | Dilute to 1X before use. The 1x solution contains 25 mM Tris, 192 mM glycine, and 0.1% SDS. |
| Tris-glycine buffer, 10X | 1st base | BUF-2020 | Dilute to 1X before use. The 1x solution contains 25 mM Tris and 192 mM glycine. |
| Ponceau S solution | Sigma Aldrich | P7170 | |
| TBS, 20X | 1st base | BUF-3030 | Dilute to 1X before use. The 1x solution contains 25 mM Tris-HCl (pH 7.5) and 150 mM NaCl. |
| Tween 20 | Sigma Aldrich | P9416 | |
| Skim Milk for immunoassay | Nacalai Tesque | 31149-75 | |
| WesternBright Sirius-femtogram HRP | Advansta | K12043 | |
| Antibody for β-actin (C4) | Santa Cruz Biotechnology | sc-47778 | Lot number: C0916 |
| MiSeq system | Illumina | SY-410-1003 | |
| NanoDrop spectrophotometer | Thermo Scientific | ND-2000 | |
| Qubit fluorometer | Thermo Scientific | Q33226 | |
| EVOS FL Cell Imaging System | Thermo Scientific | AMF4300 | |
| CRISPR plasmid: pSpCas9(BB)-2A-GFP (PX458) | Addgene | 48138 | Single vector system: The gRNA is expressed from the same plasmid. |
| CRISPR plasmid: pX601-AAV-CMV::NLS-SaCas9-NLS-3xHA-bGHpA | Addgene | 61591 | Single vector system: The gRNA is expressed from the same plasmid. |
| CRISPR plasmid: xCas9 3.7 | Addgene | 108379 | Dual vector system: The gRNA is expressed from a different plasmid. |
| CRISPR plasmid: pX330-U6-Chimeric_BB-CBh-hSpCas9 | Addgene | 42230 | Single vector system: The gRNA is expressed from the same plasmid. |
| CRISPR plasmid: hCas9 | Addgene | 41815 | Dual vector system: The gRNA is expressed from a different plasmid. |
| CRISPR plasmid: eSpCas9(1.1) | Addgene | 71814 | Single vector system: The gRNA is expressed from the same plasmid. |
| CRISPR plasmid: VP12 (SpCas9-HF1) | Addgene | 72247 | Dual vector system: The gRNA is expressed from a different plasmid. |
References
- Epinat, J. C., et al. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Research. 31 (11), 2952-2962 (2003).
- Arnould, S., et al. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. Journal of Molecular Biology. 371 (1), 49-65 (2007).
- Chapdelaine, P., Pichavant, C., Rousseau, J., Paques, F., Tremblay, J. P. Meganucleases can restore the reading frame of a mutated dystrophin. Gene Therapy. 17 (7), 846-858 (2010).
- Carroll, D. Genome engineering with zinc-finger nucleases. Genetics. 188 (4), 773-782 (2011).
- Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S., Gregory, P. D. Genome editing with engineered zinc finger nucleases. Nature Reviews Genetics. 11 (9), 636-646 (2010).
- Miller, J. C., et al. A TALE nuclease architecture for efficient genome editing. Nature Biotechnology. 29 (2), 143-148 (2011).
- Zhang, F., et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature Biotechnology. 29 (2), 149-153 (2011).
- Boch, J., et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 326 (5959), 1509-1512 (2009).
- Moscou, M. J., Bogdanove, A. J. A simple cipher governs DNA recognition by TAL effectors. Science. 326 (5959), 1501 (2009).
- Hsu, P. D., Lander, E. S., Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 157 (6), 1262-1278 (2014).
- Sander, J. D., Joung, J. K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnology. 32 (4), 347-355 (2014).
- Jinek, M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337 (6096), 816-821 (2012).
- Nishimasu, H., et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 156 (5), 935-949 (2014).
- Yamano, T., et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell. 165 (4), 949-962 (2016).
- Swarts, D. C., Mosterd, C., van Passel, M. W., Brouns, S. J. CRISPR interference directs strand specific spacer acquisition. PLoS One. 7 (4), e35888 (2012).
- Zetsche, B., et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 163 (3), 759-771 (2015).
- Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 507 (7490), 62-67 (2014).
- Hu, J. H., et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 556 (7699), 57-63 (2018).
- Kleinstiver, B. P., et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nature Biotechnology. 33 (12), 1293-1298 (2015).
- Kleinstiver, B. P., et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 523 (7561), 481-485 (2015).
- Cong, L., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 339 (6121), 819-823 (2013).
- Mali, P., et al. RNA-guided human genome engineering via Cas9. Science. 339 (6121), 823-826 (2013).
- Jinek, M., et al. RNA-programmed genome editing in human cells. Elife. 2, e00471 (2013).
- Cho, S. W., Kim, S., Kim, J. M., Kim, J. S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature Biotechnology. 31 (3), 230-232 (2013).
- Wang, Y., et al. Systematic evaluation of CRISPR-Cas systems reveals design principles for genome editing in human cells. Genome Biology. 19 (1), 62 (2018).
- Ran, F. A., et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 520 (7546), 186-191 (2015).
- Hou, Z., et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proceedings of the National Academy of Sciences U S A. 110 (39), 15644-15649 (2013).
- Kim, E., et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nature Communications. 8, 14500 (2017).
- Edraki, A., et al. A Compact, High-Accuracy Cas9 with a Dinucleotide PAM for In Vivo Genome Editing. Molecular Cell. , (2018).
- Chatterjee, P., Jakimo, N., Jacobson, J. M. Minimal PAM specificity of a highly similar SpCas9 ortholog. Science Advances. 4 (10), (2018).
- Muller, M., et al. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Therapy. 24 (3), 636-644 (2016).
- Esvelt, K. M., et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nature Methods. 10 (11), 1116-1121 (2013).
- Boratyn, G. M., et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Research. 41 (Web Server issue), W29-W33 (2013).
- Hsu, P. D., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology. 31 (9), 827-832 (2013).
- Montague, T. G., Cruz, J. M., Gagnon, J. A., Church, G. M., Valen, E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Research. 42 (Web Server issue), W401-W407 (2014).
- Heigwer, F., Kerr, G., Boutros, M. E-CRISP: fast CRISPR target site identification. Nature Methods. 11 (2), 122-123 (2014).
- Haeussler, M., et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biology. 17 (1), 148 (2016).
- Bae, S., Park, J., Kim, J. S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 30 (10), 1473-1475 (2014).
- Richardson, C. D., Ray, G. J., DeWitt, M. A., Curie, G. L., Corn, J. E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nature Biotechnology. 34 (3), 339-344 (2016).
- Richardson, C. D., Ray, G. J., Bray, N. L., Corn, J. E. Non-homologous DNA increases gene disruption efficiency by altering DNA repair outcomes. Nature Communications. 7, 12463 (2016).
- Gibson, D. G., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 6 (5), 343-345 (2009).
- Zhang, J. P., et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biology. 18 (1), 35 (2017).
- Ran, F. A., et al. Genome engineering using the CRISPR-Cas9 system. Nature Protocols. 8 (11), 2281-2308 (2013).
- Shmakov, S., et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nature Reviews Microbiology. 15 (3), 169-182 (2017).
- Moreno-Mateos, M. A., et al. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nature Communications. 8 (1), 2024 (2017).
- Lin, S., Staahl, B. T., Alla, R. K., Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 3, e04766 (2014).
- Yang, L., et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Research. 41 (19), 9049-9061 (2013).
- Watanabe, K., et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnology. 25 (6), 681-686 (2007).

