Exploring Sequence Space to Identify Binding Sites for Regulatory RNA-Binding Proteins
Summary
Sequence specificity is critical for gene regulation. Regulatory proteins that recognize specific sequences are important for gene regulation. Defining functional binding sites for such proteins is a challenging biological problem. An iterative approach for identification of a binding site for an RNA-binding protein is described here and is applicable to all RNA-binding proteins.
Abstract
Gene regulation plays an important role in all cells. Transcriptional, post-transcriptional (or RNA processing), translational, and post-translational steps are used to regulate specific genes. Sequence-specific nucleic acid-binding proteins target specific sequences to control spatial or temporal gene expression. The binding sites in nucleic acids are typically characterized by mutational analysis. However, numerous proteins of interest have no known binding site for such characterization. Here we describe an approach to identify previously unknown binding sites for RNA-binding proteins. It involves iterative selection and amplification of sequences starting with a randomized sequence pool. Following several rounds of these steps-transcription, binding, and amplification-the enriched sequences are sequenced to identify a preferred binding site(s). Success of this approach is monitored using in vitro binding assays. Subsequently, in vitro and in vivo functional assays can be used to assess the biological relevance of the selected sequences. This approach allows identification and characterization of a previously unknown binding site(s) for any RNA-binding protein for which an assay to separate protein-bound and unbound RNAs exists.
Introduction
In cell biology, gene regulation plays a central role. At one or multiple steps along the gene expression pathway, genes have the potential to be regulated. These steps include transcription (initiation, elongation, and termination) as well as splicing, polyadenylation or 3’ end formation, RNA export, mRNA translation, and decay/localization of primary transcripts. At these steps, nucleic acid-binding proteins modulate gene regulation. Identification of binding sites for such proteins is an important aspect of studying gene control. Mutational analysis and phylogenetic sequence comparison have been used to discover regulatory sequences or protein-binding sites in nucleic acids, such as promoters, splice sites, polyadenylation elements, and translational signals1,2,3,4.
Pre-mRNA splicing is an integral step during gene expression and regulation. The majority of mammalian genes, including those in humans, have introns. A large fraction of these transcripts is alternatively spliced, producing multiple mRNA and protein isoforms from the same gene or primary transcript. These isoforms have cell-specific and developmental roles in cell biology. The 5’ splice site, the branch-point, and the polypyrimidine-tract/3’ splice site are critical splicing signals that are subject to regulation. In negative regulation, an otherwise strong splice site is repressed, whereas in positive regulation an otherwise weak splice site is activated. A combination of these events produces a plethora of functionally distinct isoforms. RNA-binding proteins play key roles in these alternative splicing events.
Numerous proteins are known whose binding site(s) or RNA targets remain to be identified5, 6. Linking regulatory proteins to their downstream biological targets or sequences is often a complex process. For such proteins, identification of their target RNA or binding site is an important step in defining their biological functions. Once a binding site is identified, it can be further characterized using standard molecular and biochemical analyses.
The approach described here has two advantages. First, it can identify a previously unknown binding site for a protein of interest. Second, an added advantage of this approach is that it simultaneously allows saturation mutagenesis, which would otherwise be labor intensive to obtain comparable information about sequence requirements within the binding site. Thus, it offers a quicker, easier, and less costly tool to identify protein binding sites in RNA. Originally, this approach (SELEX or Systematic Evolution of Ligands by EXponential enrichment) was used to characterize the binding site for the bacteriophage T4 DNA polymerase (gene 43 protein), which overlaps with the ribosome binding site in its own mRNA. The binding site contains an 8-base loop sequence, representing 65,536 randomized variants for analysis7. Second, the approach was also independently used to show that specific binding sites or aptamers for different dyes can be selected from a pool of approximately 1013 sequences8. In fact, this approach has been broadly used in many different contexts to identify aptamers (RNA or DNA sequences) for binding numerous ligands, such as proteins, small molecules, and cells, and for catalysis9. As an example, an aptamer can discriminate between two xanthine derivatives, caffeine and theophylline, which differ by the presence of one methyl group in caffeine10. We have extensively used this approach (SELEX or iterative selection-amplification) to study how RNA-binding proteins function in splicing or splicing regulation11, which will be the basis for the discussion below.
The random library: We used a random library of 31 nucleotides. The length consideration for the random library was loosely based on the idea that the general splicing factor U2AF65 binds to a sequence between the branch-point sequence and the 3’ splice site. On average, the spacing between these splicing signals in metazoans is in the range of 20 to 40 nucleotides. Another protein Sex-lethal was known to bind to a poorly characterized regulatory sequence near the 3’ splice site of its target pre-mRNA, transformer. Thus, we chose a random region of 31 nucleotides, flanked by primer binding sites with restriction enzyme sites to allow for PCR amplification and attachment of the T7 RNA polymerase promoter for in vitro transcription. The theoretical library size or complexity was 431 or approximately 1018. We used a small fraction of this library to prepare our random RNA pool (~1012-1015) for the experiments described below.
Protocol
NOTE: Figure 1 provides a summary of key steps in the iterative selection-amplification (SELEX) process.
1. Generation of a random library template
- Synthesize the forward primer 5’- GTAATACGACTCACTATAGGGTGATCAGATTCTGATCCA-3’ and the reverse primer 5’- GCGACGGATCCAAGCTTCA-3’ by chemical synthesis on a DNA synthesizer.
NOTE: The primers and the random library can be synthesized commercially. - Synthesize a random library oligonucleotide template 5’- GGTGATCAGATTCTGATCCA(N1…N31)TGAAGCTTGGATCCGTCGC-3’ by chemical synthesis. Use an equimolar mixture of four phosphoramidites during synthesis for the 31 randomized positions shown above as N.
NOTE: The sequence of the library template contains 31 random nucleotides (N1 to N31) and flanking sequences for the binding of the forward and reverse primers. The forward primer includes the T7 RNA polymerase promoter sequence (underlined) for in vitro transcription and a restriction site Bcl1 (italicized) for cloning. The reverse primer contains restriction enzyme sites BamH1 and HindIII (italicized) to facilitate cloning.
2. Generation of the DNA random library pool
- Attach the T7 RNA polymerase promoter to the library by polymerase chain reaction (PCR) containing 1 µM DNA random library template, 1 µM of each primer, 20 mM Tris (pH 8.0), 1.5 mM MgCl2, 50 mM KCl, 0.1 µg/µL acetylated bovine serum albumin, 2 units of Taq polymerase, and 200 µM each of dNTPs (deoxyguanosine, deoxyadenosine, deoxycytidine, and deoxythymidine triphosphate).
- Use five cycles of denaturation, annealing, and extension steps (94 °C for 1 min, 53 °C for 1 min, and 72 °C for 1 min) of PCR followed by one cycle of extension (72 °C for 10 min).
3. Synthesis of pool 0 RNA
- Set up a 100 µL transcription reaction12. Mix T7 transcription buffer, 1 µM random library pool DNA, 5 mM dithiothreitol (DTT), 2 mM guanosine triphosphate (GTP), 1 mM each of adenosine triphosphate (ATP), cytidine triphosphate (CTP), and uridine triphosphate (UTP), and 2 units/µL T7 RNA polymerase.
NOTE: RNA can be transcribed in vitro using commercially available kits with an option of the T7 or SP6 RNA polymerase. - Incubate the above reaction mixture in a microcentrifuge tube for 2 h at 37 °C.
- Gel purify RNA in a 10% denaturing polyacrylamide gel.
- Identify location of the transcripts on the gel by staining it with methylene blue or autoradiography by including traces of radioactivity (0.5 µL or less of α-32P UTP) in the transcription reaction.
- Place the gel slice in a centrifuge tube and break into smaller pieces, for example, with a homogenizer tip. Add proteinase K (PK) buffer (100 mM Tris, pH 7.5, 150 mM NaCl, 12.5 mM EDTA, 1% Sodium dodecyl sulfate) to immerse the gel pieces. Leave the tube on a nutator from 2 h to overnight at room temperature.
- Spin in a high-speed microcentrifuge (14,000 rpm or 16,873 x g) for 5 min at room temperature to remove the gel debris and recover the buffer solution.
- Vortex the sample two times with an equal volume of phenol-chloroform and one time with chloroform.
- Mix the aqueous phase from above with one-tenth volume of sodium acetate (3.0 M, pH 5.2), 10 µg of tRNA or 20 µg of glycogen, and ethanol (2–3 volumes, stored at -20 °C). Leave the tubes at -80 °C for 1 h.
- Spin the tubes containing the solution for 5–10 min at 4 °C in a microcentrifuge (14,000 rpm or 16,873 x g). Discard the supernatant carefully. Rinse the RNA pellet with 70% ethanol and spin for 2–5 min. Aspirate ethanol carefully. Air dry the RNA pellet.
- Solubilize the RNA pellet in 50 µL of water treated with of diethyl pyrocarbonate (DEPC). Leave the sample at -20 °C for storage.
NOTE: Purify the RNA using commercially available spin columns, which are currently more commonly used to remove unincorporated radioactivity and serve as a quick and more convenient alternative for RNA purification.
CAUTION: Use an acrylic glass shield, gloves, and other precautions to protect from radioactivity.
4. Protein binding reaction and separation of bound RNA
- Carry out binding of protein and RNA in 10 mM Tris-HCl, pH 7.5 in a volume of 100 µL by adding the following ingredients to these final concentrations: 50 mM KCl, 1 mM DTT, 0.09 µg/µL bovine serum albumin, 0.5 units/µL RNasin, 0.15 µg/µL tRNA, 1 mM EDTA, and 30 µL of appropriate recombinant protein (PTB) concentration. Add RNA from the appropriate pool.
NOTE: The splicing factor U2AF65 typically binds to the polypyrimidine-tract/3’ splice sites of model introns with a binding affinity (equilibrium dissociation constant or Kd) of approximately 1–10 nM. Therefore, the first two rounds of binding used protein concentration 10-fold above the Kd for U2AF65; for SXL and PTB proteins, the starting concentration in this range was only our best guess. This ensured that desired RNA species that could bind, although lower affinity sequences also potentially bound. In rounds 3 and 4 (transcription, binding, and amplification), the protein concentration was reduced three-fold (Step 4.1). This was done to successively eliminate low affinity RNA species. - Place the tubes containing the binding reactions for about 30 min at 25 °C in a temperature block (or on ice).
- Fractionate the bound RNA from the unbound RNA for the first 4 rounds of selection-amplification using the following steps.
- Filter the sample (100 µL) at room temperature through a nitrocellulose filter attached to a vacuum manifold.
NOTE: The RNA-protein complex, but not the unbound RNA, remains on the filter. - Chop the filter with retained RNA into fragments with a sterile razor blade; insert these into a centrifuge tube. Recover RNA by tumbling the tube gently for a minimum of 3 h (or overnight) with filter pieces immersed in the proteinase K (PK) buffer.
- Deproteinize the RNA sample by vortexing it in the presence of an equal volume of phenol-chloroform (1:1) and then of chloroform. Recover the aqueous phase each time by centrifuging the sample at high speed for 5 min at room temperature.
- Mix it with sodium acetate (0.1 volume of 3.0 M, pH 5.2) and ethanol (2–3 volumes of absolute ethanol, 200 proof). Leave the tube in a -80 °C freezer for 30 min, centrifuge it at high speed for 10 min, and, following the washing and drying steps (step 3.8), solubilize the RNA in water treated with DEPC. These steps are outlined above (steps 3.6 to 3.9).
- Filter the sample (100 µL) at room temperature through a nitrocellulose filter attached to a vacuum manifold.
- Separate the protein-bound RNA fractions from the unbound fractions for the last 2 rounds (rounds 5 and 6; transcription, binding, and amplification) as follows. Reduce the protein concentration in the binding reaction (step 4.1) by further three-fold for additional selection pressure to enrich high-affinity binding sequences and preferentially remove low-affinity sequences.
- Pre-cast a native polyacrylamide gel (5% with 60:1 acrylamide:bis-acrylamide ratio) in 0.5x TBE buffer (Tris-Borate-EDTA) prior to setting up the above RNA:protein-binding (step 4.1) reaction. Electrophorese this gel in a cold room (4 °C) by applying 250 V for 15 min.
- Pipette the above RNA:protein binding reactions (step 4.1) into different wells of this gel.
NOTE: The protein is stored at -80 °C and diluted prior to use in 20 mM 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), pH 8.0, 20% glycerol, 0.2 mM ethylenediaminetetraacetic acid (EDTA), 0.05% NP-40, and 1 mM dithiothreitol (DTT). Addition of 0.5–1.0 mM protease inhibitor phenylmethane sulfonyl fluoride (PMSF) is optional. In the binding reaction, this buffer contributes about 6% glycerol, which allows direct loading of samples into the wells without the need for mixing them with a separate gel-loading buffer. - Fractionate the bound RNA from the unbound RNA using gel electrophoresis in a cold room (4 °C) at 250 V for 1 to 2 h. This process is also known as gel mobility shift assay.
NOTE: Duration of electrophoresis varies depending on features of the RNA and protein used for binding. - Expose the gel to an X-ray film and identify the location of the bound RNA using autoradiography. Cut out the gel slice with the bound RNA and insert it into a tube.
- Incubate the crushed gel slice in the PK buffer used for elution for 3 h or overnight.
- Repeat steps 3.5 to 3.9 outlined above. Briefly, vortex the eluted RNA sample vigorously first with phenol-chloroform and then with chloroform.
- Mix sodium acetate and ethanol with the aqueous phase after chloroform extraction. Following incubation in -80 °C freezer, spin at 4 °C for 5–10 min to collect the RNA pellet. Wash the RNA pellet with ethanol and air dry it by leaving the lid of the tube open. Dissolve the RNA in water treated with DEPC.
NOTE: Switching to the gel mobility shift assay for fractionation allows elimination of unwanted RNA species that might have been enriched for binding, for example, to the nitrocellulose filter here (or any matrix) used in the initial rounds for fractionation.
5. Reverse transcription and PCR amplification
- Synthesize cDNA from the dissolved RNA using reverse transcriptase and the reverse primer by incubating the 20 µL reaction (2 µL of 10x RT Buffer, 2 µL of AMV reverse transcriptase, 1 µM reverse primer, 10 µL of RNA, RNase inhibitor optional) at 42 °C for 60 min.
- Amplify the cDNA using 20–25 PCR cycles as described in step 2.1.
6. Transcription and protein binding
- Repeat the process of RNA synthesis, protein binding, separation of protein bound and unbound fraction, as described in section 3–5 above.
7. Analysis of RNA-protein interactions
- Use the gel mobility shift assay (step 4.4) or the filter binding assay (step 4.3) to determine binding affinity and specificity for selected pools or individual sequences within each pool (see step 8.3).
- Use autoradiography or a phosphor imager to detect and quantify bands in the bound fraction and unbound fraction.
8. Cloning and sequencing
- Digest the final PCR DNA product with restriction enzymes Bcl1 and HindIII for 1–2 h, ligate with the appropriately digested pGEM3 or other plasmids carrying the restriction sites from 2 h to overnight, transform the ligation product into competent bacterial cells by heat shock or electroporation using standard molecular biology procedures13.
- Grow bacteria overnight by plating the transformed cells on agar plates with Luria-Bertani (LB) medium and ampicillin (50 µg/mL) at 37 °C. Pick colonies to inoculate culture tubes containing LB liquid medium with ampicillin and grow at 37 °C in a shaking incubator overnight. Purify plasmid DNAs containing DNA inserts using a standard plasmid isolation protocol13.
NOTE: Commercial kits are available for plasmid purification. - Sequence the plasmids with DNA inserts using the dideoxy chain termination sequencing protocol, following manufacturer’s instructions for sequencing14.
NOTE: Sequencing can be performed in house or done commercially.
9. Sequence alignment
- Align sequences and obtain a consensus binding site(s) using available online alignment tools
(https://www.ebi.ac.uk/Tools/msa/).
Representative Results
The following observations demonstrate successful selection-amplification (SELEX). First, we analyzed pool 0 and the selected sequences for binding to the protein used for the iterative selection-amplification approach. Figure 2 shows that the mammalian polypyrimidine-tract binding protein (PTB) shows barely detectable binding to the pool 0 sequence but high affinity for the selected sequence pool. There was barely detectable binding to pool 0 when we used about 300-fold higher protein concentration for binding than used for the selected pool. Thus, there was at least a several hundred-fold difference in protein binding affinities between the random or starting pool and the selected pool. This observation experimentally confirms that the selection-amplification protocol described here is successful.
Second, we sequenced the selected pool and determined a consensus binding site. The consensus sequence obtained from alignment of the majority of selected sequences from the mammalian PTB-selected pool was: GCCUG(Y/G)UGCYYYYCYYYG(Y/G)CCC. This shows that we have selected unique pyrimidine-rich sequences that bind PTB11. When we performed iterative selection-amplification for the RNA-binding domain of the Drosophila PTB, we enriched CU-rich sequences interrupted by guanosines. Among the high affinity sequences that the Drosophila PTB selected was an 84% pyrimidine-rich sequence: GCUUUCCUCUGUCGCCCUUCUUCGUCCCCUG. In fact, this sequence is similar to the pyrimidine-rich sequence present in the alpha-tropomyosin intron which binds with high affinity to and is regulated by the mammalian PTB15. We have successfully used this approach repeatedly to study RNA-binding properties and functions splicing regulators and a splicing factor11, 15, 16. Table 1 shows successful examples of RNA-binding proteins for which SELEX was used to identify their preferred or consensus binding site(s).
Third, an in vitro splicing assay, which is based on alternative 3' splice site choice, shows functional relevance of distinct but overlapping RNA-binding specificities of polypyrimidine-tract binding proteins. Whereas an upstream 3' splice site is used by default, addition of the recombinant PTB leads to activation of the alternative or downstream 3' splice site (Figure 3). In contrast, addition of recombinant hnRNP C17 leads to repression of both 3' splice sites. Addition of the recombinant general splicing factor U2AF65 reverses the hnRNP C1-mediated 3' splice site repression (Figure 3) as well as the PTB-mediated effect on downstream 3' splice site activation (data not shown). A simple explanation for these effects is a direct competition between the binding of the general splice factor U2AF65 and PTB (also called hnRNP I), which preferentially binds to and represses certain 3' splices sites, or between U2AF65 and hnRNP C, which binds to and represses both 3' splice sites.
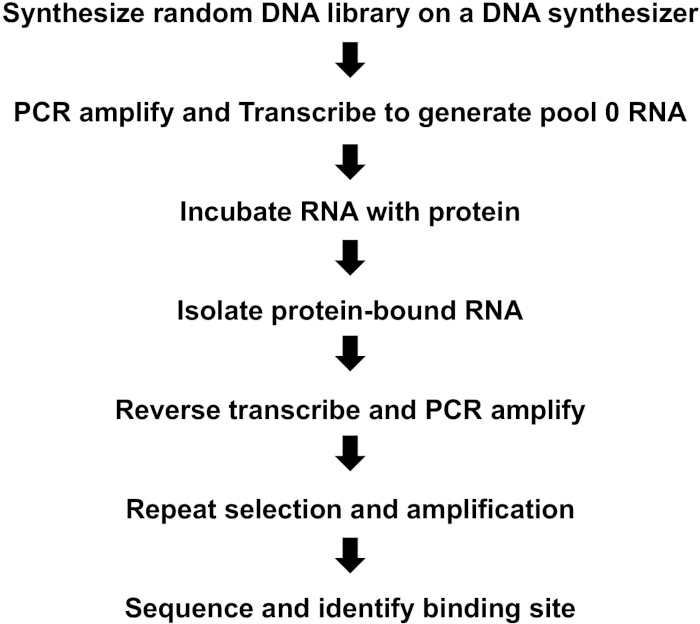
Figure 1: Summary of key steps in iterative selection-amplification process (SELEX). Please click here to view a larger version of this figure.
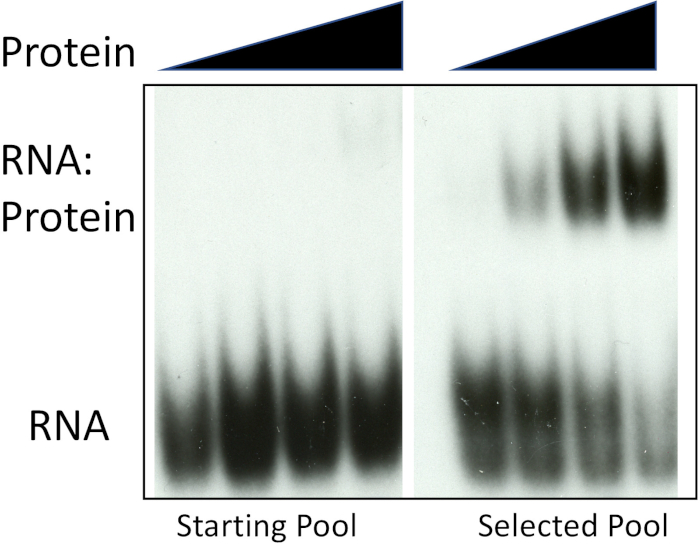
Figure 2: Enrichment of PTB-binding RNAs. Increasing concentration (filled triangles) of recombinant PTB was used with either radiolabeled pool 0 RNA or the selected pool obtained following six rounds of selection and amplification. Positions of unbound RNA and the RNA:protein complex are indicated. Please click here to view a larger version of this figure.
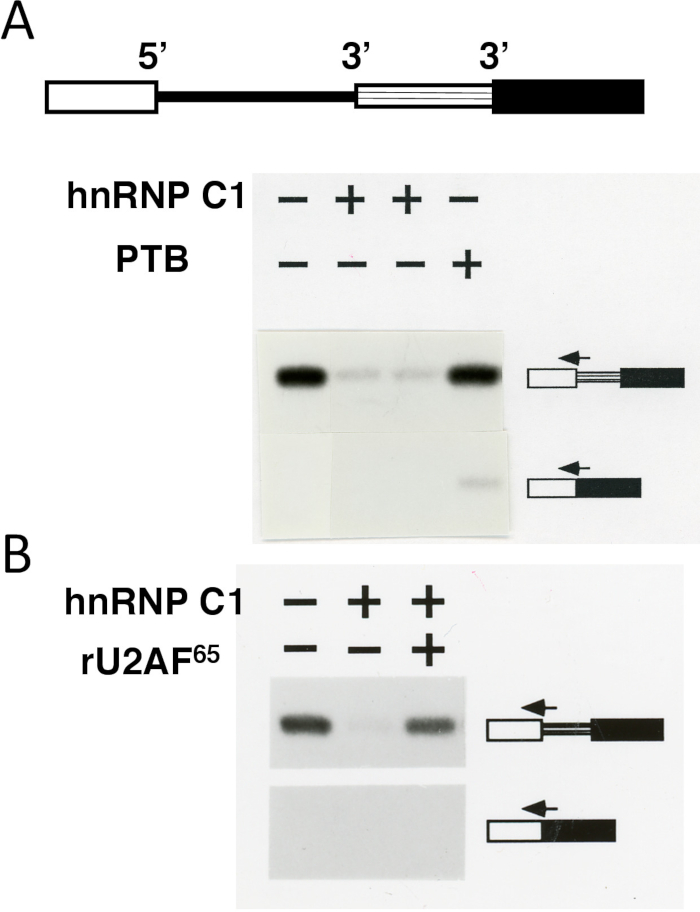
Figure 3: Splice site switching assay validates distinct binding specificities of pyrimidine-binding proteins. (A-Top) Schematics of the splicing substrate. The splicing substrate contains a 5' splice site and two alternative 3' splice sites flanking the intron. Rectangles (open, with horizontal lines, and solid) are exons and the line is an intron. (A-Bottom) hnRNP C1 represses the upstream 3' splice site (without activation of the downstream 3' splice site), whereas PTB leads to activation of the downstream 3' splice site. The splicing substrate was incubated in a HeLa cell nuclear extract.The splicing products (shown on the sides) were analyzed using a primer extension assay18 with splice-junction primers (arrows), which recognize splicing of the common 5' splice site to either the upstream or the downstream 3' splice site. (B) Recombinant U2AF65 (rU2AF65) reverses the repressive effect of hnRNP C1. Addition of the recombinant hnRNP C1, PTB, or rU2AF65 proteins to the splicing reaction is indicated by the + symbols. Please click here to view a larger version of this figure.
| Protein | Preferred sequence(s) |
| U2AF65 | U-rich containing Cs |
| SXL | U-rich containing 2-4 Gs |
| PTB | UCUUC-rich with some Gs |
| hnRNP C1 | U-rich (5-6 long) |
| CstF64 | GU-rich |
| hnRNP E1/E2 and K | C-rich |
| U2AF65/U2AF35 heterodimer | UUUYYYYUNUAGGU |
Table 1: Preferred binding sites for some RNA-binding proteins.
Discussion
Nucleic acid-binding proteins are important regulators of animal and plant development. A key requirement for the SELEX procedure is the development of an assay that can be used to separate protein-bound and unbound RNA fractions. In principle, this assay can be an in vitro binding assay such as the filter-binding assay, the gel mobility shift assay, or a matrix binding assay19 for recombinant proteins, purified proteins, or protein complexes. The assay can also be an enzymatic assay where the precursors and products (or intermediates) can be separated based on size or some other means20.
While mutagenesis has been widely used to characterize binding sites for proteins, it is laborious, and time consuming and longer sequences are not as easily amenable to saturation mutagenesis. The significance of the iterative binding and amplification approach described here is that not only does it overcome some of the above limitations, it can most importantly identify previously unknown binding sites and provide important information about nucleotide requirements at each position at the same time.
An important consideration for the success of iterative selection-amplification is binding affinity and specificity. Typically, 12 to 15 rounds of selection-amplification are employed and a sequence space of 1012 to 1015 molecules can be routinely sampled. The progress and eventual success of the selection-amplification protocol can be monitored using a binding assay or direct sequencing, which monitors affinity for or enrichment of specific sequences in intermediate pools, respectively. While the binding assay was traditionally used, advent of the next generation sequencing allows analysis of sequence enrichment in ways not possible by manual Sanger sequencing14.
A critical step in the success of SELEX is fold enrichment of the desired molecules at each step. The number of cycles required for SELEX varies and depends on several factors. For example, if fold-enrichment of desired or specific sequences is higher in each round, fewer rounds will be sufficient. However, if an assay allows a high proportion of undesired sequences in the bound pool, additional rounds will become necessary to enrich desired RNA sequences. A limitation of the technique or an unintended consequence of the need for additional cycles of selection-amplification that must be kept in mind is the possibility that it might introduce artefacts or enrich sequences that have unrelated properties such as their ability to amplify. Finally, while some applications benefit from the highest affinity binders, for other uses, a balance must be struck during the selection-amplification process between binding affinity and function because tightest binding sequences might not necessarily be the most functional sequences in biological contexts (e.g., if a sequence is recognized multiple times by different proteins during splicing).
Among the modifications and troubleshooting to improve the procedure, negative selection or counter selection can be employed to increase specificity. Similarly, use of different partitioning protocols, such as the filter binding assay followed by the gel mobility shift assay, can eliminate enrichment of unwanted sequences that bind, for example, to the nitrocellulose filter or a column matrix21. Given that proteins-nucleic acid interactions have both specific and non-specific components, buffer conditions such as salt and pH have effects on RNA-protein interactions. Moreover, use of appropriate protein concentration can have a direct effect on retention of strong, weak and non-specific binders. Selection pressure can be increased in successive rounds, for example, by including a competitor RNA, reducing protein concentration, or reducing the time of incubation. Thus, careful considerations and optimizing these parameters can impact the outcome of the SELEX protocol.
Recently, many variations or modifications of the original SELEX protocol have been developed which overcome some of the limitations mentioned above. These include high throughput-SELEX (HT-SELEX), which combines SELEX and massively parallel sequencing6, RNAcompete, which involves incubation with excess non-random RNA, pull-down of the bound RNA, fluorescent labeling of RNA, and analysis on microarrays5, RNA Bind-n-Seq, which combines RNA affinity analysis in a quantitative and high throughput fashion22, and RAPID-SELEX, which shortens the process and includes a non-amplification step23.
Chemically modified bases have been used to expand the repertoire of the RNA molecules for specific applications24. Diagnostics, therapeutics, as well as molecules with catalytic activities are among the many applications (including in medicine) of the selected molecules25. Aptamers complement antibody-based protocols and provide excellent tools whose potential, for example, in diagnostics, therapeutics, and other applications, remains to be fully exploited26,27,28. In the future, for example, clinical benefits are among the numerous desired applications, beyond what the first FDA-approved aptamer (Pegaptanib sodium) could deliver for the age-related macular degeneration. The scalable proteomic technology for protein measurements offers a step toward understanding health and diseases24.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The author thanks the National Institutes of Health for the past funding.
Materials
| Gel Electrophoresis equipment | Standard | Standard | |
| Glass Plates | Standard | Standard | |
| Nitrocellulose | Millipore | HAWP | |
| Nitrocellulose | Schleicher & Schuell | PROTRAN | |
| polyacrylamide gel solutions | Standard | Standard | |
| Proteinase K | NEB | P8107S | |
| Recombinant PTB | Laboratory Preparation | Not applicable | |
| Reverse Transcriptase | NEB | M0277S | |
| Vacuum manifold | Fisher Scientific | XX1002500 | Millipore 25mm Glass Microanalysis Vacuum Filter |
| Vacuum manifold | Millipore | XX2702552 | 1225 Sampling Vacuum Manifold |
| X-ray films | Standard | Standard |
References
- Pribnow, D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proceedings of the National Academy of Sciences of the United States of America. 72 (3), 784-788 (1975).
- Breathnach, R., Chambon, P. Organization and expression of eucaryotic split genes coding for proteins. Annual Review of Biochemistry. 50, 349-383 (1981).
- Wickens, M., Stephenson, P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3′ end formation. Science. 226 (4678), 1045-1051 (1984).
- Kozak, M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Research. 15 (20), 8125-8148 (1987).
- Ray, D., Ha, K. C. H., Nie, K., Zheng, H., Hughes, T. R., Morris, Q. D. RNAcompete methodology and application to determine sequence preferences of unconventional RNA-binding proteins. Methods. 118, 3-15 (2017).
- Jolma, A., et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Research. 20 (6), 861-873 (2010).
- Tuerk, C., Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 249 (4968), 505-510 (1990).
- Ellington, A. D., Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 346 (6287), 818-822 (1990).
- Cowperthwaite, M. C., Ellington, A. D. Bioinformatic analysis of the contribution of primer sequences to aptamer structures. Journal of Molecular Evolution. 67 (1), 95-102 (2008).
- Jenison, R. D., Gill, S. C., Pardi, A., Polisky, B. High-resolution molecular discrimination by RNA. Science. 263 (5152), 1425-1429 (1994).
- Singh, R., Valcarcel, J., Green, M. R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 268 (5214), 1173-1176 (1995).
- Milligan, J. F., Uhlenbeck, O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods in Enzymology. 180, 51-62 (1989).
- Sambrook, J., Fritsch, E. F., Maniatis, T. . Molecular Cloning. , (1989).
- Sanger, F., Nicklen, S., Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 74 (12), 5463-5467 (1977).
- Robida, M., Sridharan, V., Morgan, S., Rao, T., Singh, R. Drosophila polypyrimidine tract-binding protein is necessary for spermatid individualization. Proceedings of the National Academy of Sciences of the United States of America. , (2010).
- Banerjee, H., Rahn, A., Gawande, B., Guth, S., Valcarcel, J., Singh, R. The conserved RNA recognition motif 3 of U2 snRNA auxiliary factor (U2AF(65)) is essential in vivo but dispensable for activity in vitro. RNA. 10 (65), 240-253 (2004).
- Gorlach, M., Burd, C. G., Dreyfuss, G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem. 269 (37), 23074-23078 (1994).
- Valcarcel, J., Singh, R., Zamore, P. D., Green, M. R. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 362 (6416), 171-175 (1993).
- Wilson, C., Szostak, J. W. Isolation of a fluorophore-specific DNA aptamer with weak redox activity. Chemistry & Biology. 5 (11), 609-617 (1998).
- Joyce, G. F. Reflections of a Darwinian Engineer. Journal of Molecular Evolution. 81 (5-6), 146-149 (2015).
- McKeague, M., Derosa, M. C. Challenges and opportunities for small molecule aptamer development. Journal of Nucleic Acids. 2012, 748913 (2012).
- Lambert, N., Robertson, A., Jangi, M., McGeary, S., Sharp, P. A., Burge, C. B. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Molecular Cell. 54 (5), 887-900 (2014).
- Szeto, K., et al. RAPID-SELEX for RNA aptamers. PLoS One. 8 (12), e82667 (2013).
- Rohloff, J. C., et al. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Molecular Therapy – Nucleic Acids. 3, e201 (2014).
- Gold, L., et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 5 (12), e15004 (2010).
- Zhuo, Z., et al. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. International Journal of Molecular Sciences. 18 (10), (2017).
- Blind, M., Blank, M. Aptamer Selection Technology and Recent Advances. Molecular Therapy. Nucleic Acids. 4, e223 (2015).
- Jijakli, K., et al. The in vitro selection world. Methods. 106, 3-13 (2016).

