A Femtoliter Droplet Array for Massively Parallel Protein Synthesis from Single DNA Molecules
Summary
The overall goal of the protocol is to prepare over one million ordered, uniform, stable, and biocompatible femtoliter droplets on a 1 cm2 planar substrate that can be used for cell-free protein synthesis.
Abstract
Advances in spatial resolution and detection sensitivity of scientific instrumentation make it possible to apply small reactors for biological and chemical research. To meet the demand for high-performance microreactors, we developed a femtoliter droplet array (FemDA) device and exemplified its application in massively parallel cell-free protein synthesis (CFPS) reactions. Over one million uniform droplets were readily generated within a finger-sized area using a two-step oil-sealing protocol. Every droplet was anchored in a femtoliter microchamber composed of a hydrophilic bottom and a hydrophobic sidewall. The hybrid hydrophilic-in-hydrophobic structure and the dedicated sealing oils and surfactants are crucial for stably retaining the femtoliter aqueous solution in the femtoliter space without evaporation loss. The femtoliter configuration and the simple structure of the FemDA device allowed minimal reagent consumption. The uniform dimension of the droplet reactors made large-scale quantitative and time-course measurements convincing and reliable. The FemDA technology correlated the protein yield of the CFPS reaction with the number of DNA molecules in each droplet. We streamlined the procedures about the microfabrication of the device, the formation of the femtoliter droplets, and the acquisition and analysis of the microscopic image data. The detailed protocol with the optimized low running cost makes the FemDA technology accessible to everyone who has standard cleanroom facilities and a conventional fluorescence microscope in their own place.
Introduction
Researchers use reactors to carry out bio/chemical reactions. There are significant efforts that have been made to reduce the size of the reactor and increase the experimental throughput in order to lower the reagent consumption while improving the work efficiency. Both aspects aim to liberate researchers from a heavy workload, decreasing the cost and speeding up research and development. We have a clear historical roadmap about the development of the reactor technologies from the viewpoint of reaction volumes and throughput: single beakers/flask/test-tubes, milliliter tubes, microliter tubes, microliter 8-tube strips, microliter 96/384/1536-well plate, and microfluidic nanoliter/picoliter/femtoliter reactors1,2,3,4,5,6,7. Analogous to downsizing the feature size of transistors on integrated circuit chips in the semiconductor industry in the past decades, bio/chemical microreactors are going through volume reduction and system integration. Such small-scale tools have had a profound impact on cell-based or cell-free synthetic biology, biomanufacturing, and high-throughput prototyping and screening8,9,10,11,12. This paper describes our recent effort on the development of a unique droplet array technology and demonstrates its application in CFPS13, a fundamental technology for synthetic biology and molecular screening communities14. In particular, we intentionally provide an optimized and low-cost protocol to make the FemDA device accessible to everyone. The low-cost and easy-to-handle protocol for the miniaturized device would contribute to the educational purposes of universities and help spread the technology.
FemDA prepares femtoliter droplets at an ultrahigh density of 106 per 1 cm2 on a planar glass substrate. We coated a hydrophobic polymer, CYTOP15, on the glass substrate and selectively etched (removed) CYTOP at predefined positions to generate a microchamber array on the substrate. Thus, the resulting microchamber is composed of a hydrophobic sidewall (CYTOP) and a hydrophilic bottom (glass). When sequentially flowing water and oil over the patterned surface, the water can be trapped and sealed into the microchambers. The hydrophilic-in-hydrophobic structure is vital for repelling water outside the microchambers, isolating individual microreactors, and retaining a tiny aqueous solution inside the femtoliter space. The unique property was successfully applied for the preparation of water-in-oil droplets and lipid bilayer microcompartments16,17. Compared to the prototype device16, we first optimized the microfabrication process to realize a complete removal of the CYTOP polymer as well as a full exposure of the glass bottom. CYTOP is a special fluoropolymer featuring extremely low surface tension (19 mN/m) lower than that of conventional microreactor materials such as glass, plastics, and silicone. Its good optical, electrical, and chemical performance have already been utilized in surface treatment of microfluidic devices18,19,20,21,22,23,24. In the FemDA system, to achieve good wetting of the oil on the CYTOP surface, the surface tension of the oil must be lower than that of the solid surface25. Otherwise, the liquid oil in contact with the solid surface tends to become spherical rather than spreading over the surface. Besides, we found that some popular perfluorocarbon oils (e.g., 3M FC-40)16 and hydrofluoroether oils (e.g., 3M Novec series) can dissolve CYTOP as a result of the amorphous morphology of CYTOP, which is fatal to quantitative measurement and would be questionable in terms of cross-contamination among droplets. Fortunately, we identified a biocompatible and environmentally friendly oil exhibiting lower (< 19 mN/m) surface tension13. We also found a new surfactant that can dissolve in the selected oil and function in a low concentration (0.1%, at least 10-fold lower than previously reported popular ones26,27)13. The resulting water/oil interface can be stabilized by the surfactant. Because of the high evaporation rate of the oil, following the flush with the oil, we applied another biocompatible and environmentally friendly oil to replace the first one to seal the microchambers. We call the first oil (ASAHIKLIN AE-3000 with 0.1 wt % SURFLON S-386) the “flush oil” and the second oil (Fomblin Y25) the “sealing oil,” respectively.
The two-step oil-sealing strategy can realize a robust formation of the femtoliter droplet array within minutes and without sophisticated instrumentation. Due to the evaporation problem, it has been considered challenging to generate microreactors smaller than picoliter volumes28. FemDA addressed this issue by systematically optimizing the materials and processes used for the preparation of microreactors/droplets. Several noteworthy features of the resulting droplets include the high uniformity (or monodispersity), stability, and biocompatibility at the femtoliter scale. The coefficient of variation (CV) of the droplet volume is only 3% (without vignetting correction for the microscopic images), the smallest CV among droplet platforms in the world, which ensures a highly parallel and quantitative measurement. The femtoliter droplet is stable for at least 24 hours without cross-contamination among droplets at room temperature, which is valuable for a reliable time-course measurement. Regarding the biocompatibility, we succeeded in synthesizing various proteins from a single-copy template DNA in the femtoliter droplet, which had previously been considered difficult or inefficient29,30. It would be worthy of elucidating why some proteins capable of being synthesized in the FemDA cannot be synthesized in other droplet systems. FemDA was not merely a technical advancement, but also realized an unprecedentedly quantitative measurement that can correlate the protein yield (as reflected by the fluorescence intensity of the droplet) to the number of template DNA molecules in each droplet. As a result, the histogram of the fluorescence intensity of droplets from FemDA-based CFPS showed a discrete distribution that can be nicely fitted by a sum of Gaussian distributions of equal peak-to-peak intervals. Moreover, the probability of occurrence of droplets containing different numbers of DNA molecules was a perfect fit to a Poisson distribution31. Thus, the different protein yield in each droplet can be normalized based on the discrete distribution. This critical feature allows us to separate the enzymatic activity information from the apparent intensity, that has not been available with other microreactor platforms yet. Existing microfluidic cell/droplet sorting systems are skilled in fully automatic sorting and good at concentrating samples but sometimes can only output a relatively broad or long-tailed histogram in the analytical aspect32,33. Our highly quantitative and biocompatible FemDA system sets a new benchmark and a high analytical standard in the field of microreactor development.
The oils and surfactants that could be used for the preparation of droplets are still very limited34. The combination of ASAHIKLIN AE-3000 and SURFLON S-386 established in FemDA is a new member of the growing arsenal of the physiochemical interface between the aqueous phase and the oil phase13. The new interface in FemDA is physically stable, chemically inert, and biologically compatible with the complex transcription, translation, and post-translational modification machinery for many sorts of proteins13. It would be rather attractive to find a protein that cannot be synthesized in the droplet settings instead. Besides, the cost saving of reagents is more evident in the femtoliter droplet system than that in nanoliter and picoliter reactor systems35,36. In particular, there would often be a large dead volume, which is mainly caused by tubing or external supplies, in microfluidic droplet generation systems but not in our FemDA. The array format is also favored by repeated and detailed microscopic characterization (similar to so-called high-content analysis) for every single reactor37, rather than only a single snapshot for a fast-moving object. The femtoliter scale enabled the integration of over one million reactors on a finger-sized area, while the same number of nanoliter reactors (if it exists) requires over square meter area, which would be undoubtedly impractical to manufacture or use such system.
Protocol
1. Microfabrication of the femtoliter microchamber array substrate
NOTE: Conduct the following microfabrication experiment in a cleanroom. Wear gloves and a cleanroom suit before entering the cleanroom.
- Cleaning cover glass substrate
- Set the cover glass on a cover glass staining rack. Sonicate the cover glass in 8 M sodium hydroxide (NaOH) for 15 min at room temperature (RT).
CAUTION: NaOH in high concentrations is highly dangerous to skin and eye. Gently handle it without any splash. - Take the rack out of the NaOH solution and rinse the cover glass using water for ten times. Keep the cover glass in the pure water.
NOTE: Discard the alkaline waste to the designated tank. The NaOH solution can be recycled to the original bottle and used for up to five times. - Dry each piece of cover glass with an air blow gun.
- Bake the dried cover glass on a hot plate at 200 °C for 5 min. Keep the orientation of the upper side of the glass substrate consistent during the following handling processes.
- Set the cover glass on a cover glass staining rack. Sonicate the cover glass in 8 M sodium hydroxide (NaOH) for 15 min at room temperature (RT).
- Silanization of the glass surface
- Reset the cleaned cover glass on a dry staining rack.
- Add 150 mL of ethanol into a PTFE beaker. Use a needle (22 G x 70 mm) equipped 1 mL syringe to draw 0.075 mL of (3-aminopropyl)triethoxysilane and immediately inject it to the ethanol (i.e., 0.05 vol %). Pull and push the syringe several times to rinse the inside of the syringe. Stir the solution with the needle to make it homogeneous.
NOTE: (3-aminopropyl)triethoxysilane can be stored at room temperature and 1 atm pressure for two years with a tight sealing. Absolute ethanol should be tightly sealed after use. We do not recommend using any expired reagents. - Incubate the cover glass in the 0.05 vol % aminosilane solution for 1 h at RT.
- Rinse the cover glass using pure water for five times. Keep the cover glass in the pure water.
NOTE: Discard the waste to the designated tank. - Dry each piece of cover glass with the air blow gun.
- Place all the dried cover glass on aluminum foil. Bake the cover glass on the hot plate at 80 °C for 5 min.
- Spin-coating CYTOP perfluoropolymer
- Place the cover glass on a customized vacuum chuck of a spin coater (Figure 1A). Turn on the vacuum switch to fix the glass substrate.
NOTE: The design of the vacuum chuck is crucial for a uniform coating of viscous polymer on the rectangular (24 mm × 32 mm) and thin (0.13-0.17 mm) substrate (Figure 1A). We found that a rectangular sample stage with multiple holes (48 holes, each with 1 mm diameter) connecting to the vacuum channel worked better than the round stage with a single central hole did. - Drop 70-90 µL of type-A CYTOP polymer at the center of the glass substrate. Immediately spin-coat the polymer (Figure 1B).
NOTE: The spin-coating condition (3,400 rpm, 30 s) provides 3 μm thickness. Use the micropipette designed for viscous liquid. Hold the micropipette upright to make the polymer drop near-perfectly circle on the substrate. An air bubble is often generated inside the pipette tip as the plunger moves downward. Do not drop the last drop of CYTOP with the bubble. - Pick up the coated glass with fingers holding corners of the glass substrate, and place it on aluminum foil.
- Repeat steps 1.3.1-1.3.3 to spin-coat all the remaining pieces of the cover glass.
- Bake the coated glass at 80 °C for 30 min and then at 200 °C for 1 h.
NOTE: This processing fully cures the CYTOP and covalently bond the CYTOP to the glass surface. Cover the baking area with a lid made of aluminum foil to avoid potential dust during the long-time baking process if needed. - Remove the CYTOP-coated cover glass from the hotplate and cool down to RT. Carefully pick up each piece of the coated glass and inspect it to see if it is properly coated. Discard the substrate exhibiting inhomogeneous coating.
NOTE: The surface of a nicely coated CYTOP should look flat without irregular rainbow-like patterns (Figure 1C). Visual inspection from a proper angle can rapidly judge the flatness as well as the coating quality. The protocol can be paused here.
- Place the cover glass on a customized vacuum chuck of a spin coater (Figure 1A). Turn on the vacuum switch to fix the glass substrate.
- Spin-coating photoresist
- Place the CYTOP-coated cover glass on the vacuum chuck (see step 1.3.1) of the spin coater. Turn on the vacuum switch to fix the cover glass.
- Drop 0.2-0.3 mL of photoresist at the center of the substrate. Immediately spin-coat the photoresist at 6,000 rpm for 60 s.
NOTE: Do not drop the last drop of the photoresist with bubbles. If the spin coater supports higher spin speed, the spin speed could be up to 7,500 rpm to achieve a thinner coating. The low surface energy of CYTOP prevents most of the photoresists from being adhered to its surface. If AZ P4903 photoresist is not available, AZ P4620 photoresist is a good alternative. - Pick up the coated substrate with two fingers holding corners of the substrate. Wipe off the excess photoresist near the substrate edge (often called as edge bead removal or EBR) using an ethanol-soaked clean wiper (Figure 1D). Place the substrate on the aluminum foil.
NOTE: Acetone is NOT recommended for EBR. - Repeat steps 1.4.1-1.4.3 to spin-coat the photoresist for the remaining pieces of CYTOP-coated cover glass. Pick up every piece of the coated glass and inspect it to see if it is properly coated.
NOTE: The surface of a nicely coated photoresist should look flat without irregular patterns (Figure 1D). Visual inspection from a proper angle can rapidly judge the flatness as well as the coating quality. The substrate exhibiting inhomogeneous coating can be recycled through washing with acetone, 2-propanol, and H2O in sequence, and reused. - Bake the coated substrate at 110 °C for 5 min on the hotplate.
- Remove the coated substrate from the hotplate and cool down to RT. Let stand for 30 min at a relative humidity of 40%-60% for rehydration of the photoresist.
NOTE: H2O is necessary for the following photochemical reaction. The baked photoresist lost the H2O content inside the photoresist. The rehydration process allows H2O to be absorbed from the air. Relative humidity lower than 20% or higher than 80% is not suitable for the rehydration process.
- Photolithography
- During the waiting time of rehydration (see step 1.4.6), start up the mask aligner. Warm up the light source. Load the chrome photomask.
NOTE: Follow the instruction manual to operate the mask aligner. The photomask can be fabricated via electron beam (EB) lithography if the EB facility is available. Alternatively, the photomask can be outsourced to a local company. - Load the rehydrated substrate (after finishing step 1.4.6) on the chuck (Figure 1E).
- Set exposure parameters. Expose the substrate for 25 s with a UV intensity of 13 mW/cm2 (h-line) in the vacuum-contact mode. Then unload the substrate and set it on the cover glass staining rack (the same rack used in step 1.1.1).
- Repeat steps 1.5.2-1.5.3 to expose the remaining pieces of the rehydrated substrate.
- During the waiting time of rehydration (see step 1.4.6), start up the mask aligner. Warm up the light source. Load the chrome photomask.
- Development
- Develop the substrate for 5 min to dissolve the exposed photoresist (Figure 1F). During which, gently shake the rack in the developer once at the timepoint of 4 minute.
NOTE: The developer was AZ 300 MIF. Alternative alkaline developers (e.g., AZ 400 K) are generally applicable for the development but should require optimization of development conditions (e.g., time, temperature, agitation). Do NOT use glass beakers as the developer container. - Rinse the substrate using pure water for ten times. Keep the cover glass in the pure water.
NOTE: Discard the developer to the designated tank. - Thoroughly dry every piece of substrate with the air blow gun.
- Develop the substrate for 5 min to dissolve the exposed photoresist (Figure 1F). During which, gently shake the rack in the developer once at the timepoint of 4 minute.
- Reactive-ion etching
- Place the dried substrate (from step 1.6.3) in the reaction chamber of the reactive-ion etching (RIE) machine.
NOTE: According to the maintenance rule of the facility, the RIE machine can be either always on or shut down every time. If the switch was off, turn on the switch according to the manual. - Etch the photoresist-uncovered CYTOP with the O2 plasma (Figure 1G).
NOTE: The RIE condition was as follows. O2 gas flow rate: 50 sccm; chamber pressure: 10.0 Pa; RF power: 50 W; etching time: 27 min. The etching time was optimized for completely etching 3 μm thickness of CYTOP. - Pick up every piece of substrate from the reaction chamber using the tweezer and place them on the cover glass staining rack.
NOTE: According to the maintenance rule of the facility, the reaction chamber may require a short O2 plasma cleaning process (e.g., etching time: 5 min) to keep the reaction chamber clean after every run.
- Place the dried substrate (from step 1.6.3) in the reaction chamber of the reactive-ion etching (RIE) machine.
- Removing photoresist and cleaning
- Sonicate the substrate in acetone for 5 min at RT to dissolve the remaining photoresist.
- Sonicate the substrate in 2-propanol for 5 min at RT.
- Rinse the substrate using pure water for five times. Sonicate the substrate for 5 min at RT. Rinse the sample using pure water for another five times. Keep the substrate in pure water.
- Dry every piece of substrate with the air blow gun (Figure 1H).
NOTE: The substrate can be stored in a plastic Petri dish. The fabricated substrate is structurally stable at RT for at least one year. The protocol can be paused here. - Characterize the surface profile of the as-prepared substrate by three-dimensional (3D) laser scanning confocal microscopy, white light interferometry (or coherence scanning interferometry), or scanning electron microscopy. Use the respective software to measure the diameter and depth of the resulting microchambers.
NOTE: The sample can be reused for other experiments after the characterization with the non-contact and non-destructive 3D laser scanning confocal microscope or white light interferometer. The protocol can be paused here.
2. Preparation of polydimethylsiloxane (PDMS) microchannel
NOTE: Do NOT wear latex gloves to handle PDMS. Instead, wear polyethylene (PE) gloves.
- Making the microchannel master
- Cut a double-coated adhesive Kapton film tape into a defined (3 mm x 19 mm) microchannel shape using a desktop cutter.
NOTE: The Kapton tape was No. 7602 #25 (Teraoka Seisakusho), resulting in a channel height of 135 μm. The STIKA desktop cutter uses a plugin (available from Roland website: https://www.rolanddga.com) of Adobe Illustrator to run the cutting according to the drawing in the Adobe Illustrator file. The protocol can be paused here. - Stick every cut Kapton tape on the flat bottom of a Petri dish. The standard 90 mm Petri dish can accommodate 12 pieces of the tape.
NOTE: The relief structure serves as a master for the replica molding of PDMS. Press the tape surface gently with a tweezer if there are any air bubbles sandwiched between the tape and the Petri dish substrate. Alternatively, classical soft-lithography can be applied to prepare the master on a silicon wafer38. The protocol can be paused here.
- Cut a double-coated adhesive Kapton film tape into a defined (3 mm x 19 mm) microchannel shape using a desktop cutter.
- Curing PDMS resin in the tape-patterned Petri dish
- Wear new PE gloves, and transfer 2.5 g of curing agent of SYLGARD 184 silicone elastomer using a disposable plastic pipette into the specified plastic beaker (Figure 2A).
- Change into new gloves, and transfer 25 g of the prepolymer base of SYLGARD 184 silicone elastomer using a disposable 50 mL syringe into the above plastic beaker.
NOTE: Change the gloves herein to prevent the potential cross-contamination of the curing agent to the base. - Use a deaeration mixer to mix and deaerate the mixture (program: 3 min mixing followed by 2 min deaeration).
NOTE: If the deaeration mixer is not available, a manually agitated (for about 15 min) mixture can be degassed in a vacuum chamber. - Pour the PDMS mixture into the tape-patterned Petri dish (Figure 2B). Set the Petri dish in a mini vacuum chamber and deaerate the PDMS mixture for 1-3 h (Figure 2C).
NOTE: If any air remains in the PDMS mixture after 1 h, take the Petri dish out of the vacuum chamber, break the air bubble using an air blower, and then put the Petri dish back in the vacuum chamber and continue the deaeration process. - Place the Petri dish in an oven at 60 °C overnight to cure the PDMS (Figure 2D).
NOTE: Although increasing the temperature can shorten the curing time, the maximum temperature that can be tolerated by the polystyrene Petri dish without physical deformation is about 60 °C.
- Replica molding of PDMS channel
- Peel off the cured PDMS elastomer from the Petri dish (Figure 2E).
- Cut off every piece of PDMS channel blocks using a flat-cable cutter (Figure 2F).
- Place the PDMS elastomer on a cutting mat facing channel side up. Punch a hole at each end of the channel using a biopsy punch (Figure 2G).
- Clean the surface of the PDMS channel with Scotch tape. Then wrap the PDMS resin in another piece of Scotch tape to keep it clean before use. Store the prepared PDMS channel in a clean Petri dish.
NOTE: The protocol can be paused here.
3. Assembly of femtoliter microchamber array device
- Place some pieces of water-soaked clean wipers along the inside wall of a Petri dish to make a simplified humidifying chamber. Place the PDMS resin covered by the Scotch tape in the humidifying chamber and seal the chamber using paraffin film. Incubate for at least 3 h but no longer than a day.
NOTE: PDMS is a porous material that allows gas to pass across the resin39. The porous structure of PDMS leads to an absorption of water molecules from the surroundings until reaching an equilibrium. This pre-treatment fills the pores of PDMS with water vapor and can significantly repress the evaporation of aqueous droplets adjacent to the edge of the PDMS channel. - Take the Scotch tape off the PDMS resin. Position the PDMS channel on the microchamber array area of the substrate (Figure 2H).
NOTE: The PDMS resin reversibly adheres to the CYTOP surface. Press the PDMS block gently with a tweezer to remove any air bubbles in between the PDMS resin and the substrate if existed. - Insert a 200 μL non-filtered pipette tip to one (as the outlet) of the holes of the PDMS channel.
NOTE: The adhesion strength between PDMS and CYTOP is limited. To avoid physical deformation of the PDMS resin as well as detachment of the PDMS channel off the surface, do not insert the pipette tip too deep.
4. Loading reaction solution to the assembled device
- Put an aluminum microtube stand on the ice. Prechill the flush oil (ASAHIKLIN AE-3000 oil mixed with 0.1 wt % SURFLON S-386 surfactant) in a 1.5 mL microtube on the aluminum stand.
- Put another aluminum block on ice.
- Preparation of CFPS reaction solution
- Prepare the CFPS reaction solution in a PCR tube. Mix every aliquot component of the CFPS kit, a diluted template DNA (as exemplified herein by fluorescent protein mNeonGreen40 and Escherichia coli alkaline phosphatase41) solution, and other necessary components according to the specific needs (e.g., RNase inhibitor, fluorogenic substrate, chaperone).
NOTE: The template DNA used for CFPS must be prepared according to the instruction of the corresponding CFPS kit. This demonstration applied a T7-based expression system42. Because the reagent consumption in the FemDA device is quite small, 10 μL is large enough to fill the entire PDMS channel. - For mNeonGreen fluorescent protein synthesis, mix the components in the following order: add 2.7 μL of nuclease-free H2O to a 6 μL aliquot of solution A (from the CFPS kit); add 0.3 μL of RNase inhibitor; add 1.5 μL of diluted template DNA solution; briefly mix and transfer the mixture to a 4.5 μL aliquot of solution B (from the CFPS kit).
NOTE: The total volume is 15 μL. As the quantity of every aliquot is proportional to the total volume of the final mixture, a total volume less than 10 μL may result in a far too small volume (< 0.2 μL) of some component aliquot. - For alkaline phosphatase synthesis, mix the components in the following order: add 1.2 μL of H2O to 6 µL of solution A; add 0.3 µL of RNase inhibitor; add 0.6 μL of disulfide bond enhancer 1 and 2 (from a kit); add 0.3 μL of 5 mM 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP); add 1.5 μL of DNA solution; briefly mix and transfer the mixture to 4.5 μL of solution B.
NOTE: In the assay of alkaline phosphatase, the fluorogenic substrate DiFMUP can produce highly fluorescent 6,8-difluoro-7-hydroxy-4-methylcoumarin (DiFMU) upon enzymatic cleavage of the terminal phosphate group.
- Prepare the CFPS reaction solution in a PCR tube. Mix every aliquot component of the CFPS kit, a diluted template DNA (as exemplified herein by fluorescent protein mNeonGreen40 and Escherichia coli alkaline phosphatase41) solution, and other necessary components according to the specific needs (e.g., RNase inhibitor, fluorogenic substrate, chaperone).
- Draw up 10 μL of the mixture using a 200 μL non-filtered pipette tip. Insert the pipette tip to the inlet hole (refer to step 3.3) of the PDMS channel. Push down on the plunger to inject the solution to the channel until it overflows from the outlet (where another pipette tip has already been inserted at step 3.3).
- Separate the pipette from the pipette tip and keep these two pipette tips still inserted to the inlet and outlet.
NOTE: Avoid generating air bubbles during the pipetting. Do not leave the thumb from the plunger until separating the pipette from the pipette tip to prevent the backflow of the solution inside the PDMS channel.
5. Generating femtoliter droplet array (FemDA) for CFPS
- Transfer the whole set of the assembled device to the pre-chilled aluminum block (see step 4.2). Carefully confirm that the microchamber array area quickly turns from translucent (Figure 2I) to transparent (Figure 2J).
NOTE: The CFPS reaction solution cannot straightly enter into the microchambers. The solubility of air in water is in inverse proportion to the temperature. The trapped air will dissolve into the solution after the device was moved from RT to the low temperature. The transparency change is a convenient visual indicator to judge whether the solution enters into the microchambers. - Draw 30 μL of the pre-chilled flush oil (see step 4.1), and immediately transfer the oil into the inlet-inserted pipette tip from its upper opening. The flush oil moves into the channel and extrudes the excess reaction solution located outside the microchambers. Every microchamber is isolated by the flush oil.
NOTE: The moving interface between the reaction solution and the flush oil is visible, which helps the people to observe the movement of the fluid inside the channel. The extruded solution can be collected into the previous tube, stored at 4 °C, and reused for another FemDA device or a parallel bulk reaction (in the microtube or a microtiter plate). - Pre-draw 30 μL of sealing oil (Fomblin Y25) to a 200 μL filtered pipette tip. Hang the pipette upright somewhere.
- Simultaneously remove the two inserted pipette tips (see steps 3.3 and 4.4) from the device. Immediately move the device from the aluminum block to parafilm.
NOTE: Use a tweezer to fix (but keep away from the channel area) the device on the aluminum block during the period of removing the pipette tips. - Immediately insert the ready pipette tip containing the sealing oil (see step 5.3) into the inlet of the PDMS channel, and inject the oil into the channel until it overflows from the outlet.
NOTE: Carry out this step as soon as moving the device to RT. To prevent the backflow inside the channel, do not leave the thumb from the plunger until the sealing oil overflows from the outlet. The flush oil evaporates immediately after moving out of the outlet of the channel, while the following sealing oil accumulates in the outlet because of its extremely low evaporation loss. - Separate the pipette from the pipette tip and then remove the pipette tip from the device. Clean the PDMS surface using a piece of the clean wiper and then apply a new drop of sealing oil to the inlet and outlet, respectively. The femtoliter droplets are sealed in individual microchambers by the oil.
NOTE: Use a tweezer to fix (but keep away from the channel area) the PDMS channel on the substrate during the period of removing the micropipette tip. - Wipe the moisture accumulated on the lower surface of the cover glass. The as-prepared FemDA device is now ready for incubation and microscopy imaging.
6. Microscopy imaging
- Start up an inverted fluorescence microscope. Wait for the stabilization (cooling) of the camera. Use a 60x or 100x oil immersion objective lens. Set the FemDA device on the motorized stage. Set the imaging parameters.
NOTE: The imaging parameters include the file path, imaging channels, filters, exposure times (in general, 100 ms is enough; shorter or longer time is also possible according to the specification of the camera), and light intensity. - Find the focal plane. Adjust the z-axis position of the objective lens and fix the focal plane using an internal autofocus system. Set the region of interest (ROI; i.e., x, y-axis) to be imaged.
NOTE: The major microscope manufacturers (Nikon, Olympus, Zeiss, Leica) all provide the option to integrate the autofocus system with the microscope. This autofocus system is necessary for keeping the focal plane constant during the automatic multi-area imaging. The ROIs cover the FemDA area in the PDMS channel. - Capture the fluorescence images. Capture a single frame of a defocused bright-field (BF) image for every ROI. Do NOT change the order and the coordinates of the ROIs when imaging the different channels.
7. Image data analysis
NOTE: Analyze the image data using a homemade plugin (named “FemDA”) based on Fiji (http://fiji.sc) to extract the fluorescence intensity of each droplet43. Install the correct version of Fiji according to the operating system. Fiji supports most image file formats (e.g., the nd2 file from Nikon microscope, czi file from Zeiss microscope) using a build-in plugin “Bio-Formats.”
- Plugin installation
- Copy and paste the plugin file (which is available upon inquiry to the corresponding author or via the following institutional link: https://fbox.jamstec.go.jp/public/FcsQwAsMKIqA9j0B2MJwLUMeHGsxp09hAkAFdVhQIDkZ) into Fiji’s “plugins” folder.
- Start the Fiji software; the plugin “FemDA” can be found in the drop-down menu “Plugins” of Fiji.
- Image data preprocessing
- Open the defocused BF image (see step 6.3). Click Process | Subtract Background… to remove smooth continuous backgrounds of every frame of the image data (Figure 4B). Click Process | Filters | Median to reduce the noise of every frame of the image data (Figure 4C).
- Click Image | Adjust | Threshold to check and separate the foreground (indicating the microchambers) from the background (indicating the area outside the microchambers) of every frame of the image data (magnified inserts in Figure 4A-C).
NOTE: Select a proper algorithm from the drop-down list in the Threshold window so that only the microchamber areas, as well as the edges of every microchamber, can be identified and highlighted as the foreground. In particular, most of the highlighted edges must be continuous without gaps.
- Droplet coordinate determination
- Click Plugins | FemDA | FemDA Analysis to open the graphic user interface (GUI) of the customized plugin (upper half of Figure 4D).
- Input the approximate minimum and maximum numbers of pixels of a single microchamber. Input the expected minimum and maximum circularity (0: arbitrary shape; 1: perfect circle) of the microchambers. Input the frame number of the start frame and the end frame.
- Click Generate ROI on the GUI to detect every microchamber, and the successfully detected ROIs are shown in a popup window ROITable.
- Go back to the window FemDA Analysis and click the button Apply ROI mask to check if the microchambers over the frames were properly detected (lower left of Figure 4D). If no, modify some of them and repeat clicking Generate ROI and Apply ROI mask. If yes, go to the next step.
NOTE: There may be a very few microchambers that cannot be well identified as the ROI (see the lower half of Figure 4D) by any algorithm of the Threshold because of the insufficient contrast between the foreground and the background. It is not problematic in the statistical viewpoint.
- Fluorescence intensity extraction
- Open the fluorescence image and bring it to the front. Click Apply ROI mask again to apply the determined ROIs onto the fluorescence image (lower right of Figure 4D).
- Select either One-Shot for analyzing an end-point image (as exemplified with the mNeonGreen data) or Time-Lapse for analyzing a time-course data (as exemplified with the alkaline phosphatase data).
- Input 100 in the box Number of top pixels means that only the top 100 pixels of each ROI will be used to calculate the mean intensity of the corresponding droplet. If input 0 in the Number of top pixels, all pixels of each ROI will be used to calculate the mean intensity of the corresponding droplet.
NOTE: There may be a drift of several pixels between the BF and the fluorescence images, which means some pixels within the ROIs may belong to the background of the fluorescence image. Hence, the plugin provides an option to specify the number of top intensity-ranked pixels for the calculation of the mean intensity of every droplet. - For the analysis of the end-point image (i.e., select One-Shot at step 7.4.2), click button Measure intensity to calculate the mean intensities of all detected droplets. The ROITable is updated with the new data from the fluorescence image. Meanwhile, a histogram is displayed in a new popup window Histogram (Figure 4E).
NOTE: Changing the bin size in the box Bin Number can optimize the display of the histogram. A fitting to a sum of Gaussian distributions can be accomplished in the developed GUI. The popup Log window of Fiji displays the fitting results. Alternatively, export the data by clicking the button save as text and carry out various statistical analyses with other software (Figure 4F, G). - For the analysis of the time-course image (i.e., select Time-Lapse at step 7.4.2), the popup window is Intensity Time Course instead of Histogram, which contains a histogram corresponding to a specific time-point and a time-intensity plot (Figure 5). The textual data can also be exported using the File menu of the popup window.
Representative Results
The microfabrication process consists of substrate cleaning, surface functionalization, CYTOP coating, photolithography, dry etching, photoresist stripping, and final cleaning. Importantly, the presented protocol allowed complete removal of the hydrophobic CYTOP polymer inside the microchambers Figure 3A), producing a highly parallel hydrophilic-in-hydrophobic structure on a standard cover glass substrate. With the aid of the oil sealing protocol, the uniform dimension of the resulting droplets was verified by encapsulating fluorescent solution in the microchambers (Figure 3B). The fluorescence intensity extracted using the developed software is a good indicator of the droplet size. The CV of the fluorescence intensity, 3%, reflected the narrow distribution of the droplet size over the entire array (Figure 3C). The reconstructed 3D image from confocal microscopy also showed the consistent droplet volume over time (Figure 3D, Supplementary Movie 1)44. In comparison, a widely used FC-40 oil generated the droplets exhibiting a severalfold difference in the fluorescence intensity45. The formed droplets in FemDA were stable at RT for at least 24 hours (Figure 3E). The high quality of the droplets eventually formed the basis of quantitative measurement.
High-throughput data necessitate high-throughput data-analyzing tools46,47. The developed plugin greatly simplified and speeded up the image data analysis. Based on the theory of digital image processing48, the defocused BF image can be binarized and used to provide the coordinate information of every droplet after background correction and noise reduction (Figure 4A-C). This idea made the precise localization of dark droplets possible.
The fluorescent protein mNeonGreen was synthesized in FemDA. Template DNA with a concentration of 0.05 molecules per droplet was randomly distributed into each droplet to initiate the protein synthesis with coupled cell-free transcription and translation reactions. Because the average number of DNA molecules per droplet was smaller than 1, some droplets contained zero template DNA, while others contained one or more DNA molecules. After 6 hours of incubation at RT, the end-point image was captured using the microscope. The stack image data was analyzed by the developed software with the aid of the concurrent defocused BF image (Figure 4A-E). The fluorescence intensity of each droplet is a measure of the protein yield in the corresponding droplet. The histogram of the fluorescence intensities showed a discrete distribution and was well fitted by a sum of Gaussian distributions of equal peak-to-peak intervals (Figure 4F), which strongly suggested an occupancy of different numbers of DNA molecules per droplet. Similar to the repeated coin-flipping game, the number of independent random events that occur is mathematically described by the Poisson distribution. The probability of occurrence of droplets containing different numbers (up to 3 in this example) of DNA molecules was a perfect fit to a Poisson distribution (P = e-λ • λk / k!, where λ is the expected average number of DNA molecules per droplet and k is the actual number of DNA molecules in a droplet) with an average of 0.05 DNA molecules per droplet (Figure 4G)31, as expected for a random distribution of DNA molecules. Because the loading concentration of the template DNA solution was the same as the final fitted λ (i.e., 0.05), the CFPS reaction efficiency in our FemDA was 100% (or near 100%). In other words, a single DNA molecule is enough to trigger the CFPS reaction in the femtoliter droplet efficiently.
The CFPS reaction of alkaline phosphatase was recorded every 5 minutes. The developed software also supports analyzing the time-course data (Figure 5). The coupled fluorogenic reaction showed a similar discrete distribution of the fluorescence intensity of the droplets at earlier time-points. The histogram results also verified an occupancy of different numbers of DNA molecules in the droplet. The fluorescence intensity eventually converged to a value along with the gradual depletion of the fluorogenic substrate DiFMUP.
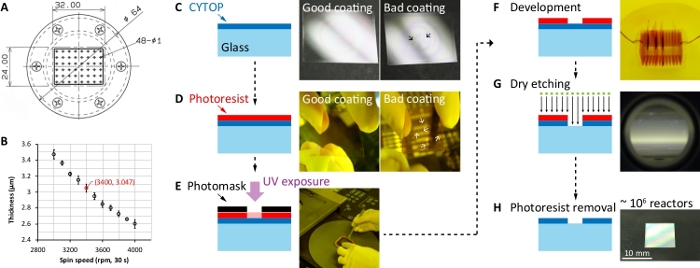
Figure 1: Microfabrication process of the ultrahigh-density microchamber array substrate. (A) Technical drawing of the customized vacuum chuck. Unit: mm. (B) CYTOP film thickness vs. spin speed data. (C) Spin-coating of the perfluoropolymer CYTOP on a silanized glass substrate. The photographs showed a good example of homogenous coating and a bad example of inhomogeneous coating, respectively. The black arrow indicated the specific position of the inhomogeneous CYTOP film. (D) Spin-coating of photoresist on the CYTOP-coated substrate. After the spin-coating, the photoresist near the substrate edge must be removed using an ethanol-soaked clean wiper (middle photograph). The photographs showed a good example of homogenous coating and a bad example of inhomogeneous coating, respectively. The white arrow indicated the specific position of the inhomogeneous photoresist film. (E) Exposure of the coated photoresist using a mask aligner. (F) Development of the exposed photoresist in a developer. The exposed part of the photoresist is soluble in the developer solution. (G) Reactive-ion etching of CYTOP. The uncovered CYTOP was removed by O2 plasma. (H) Removal of the photoresist mask. The photoresist was removed by acetone. The substrate was cleaned using 2-propanol and H2O. Please click here to view a larger version of this figure.
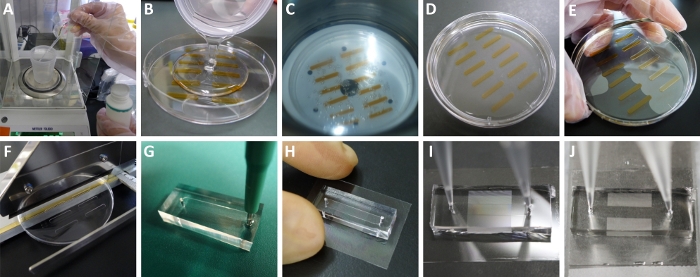
Figure 2: Preparation and use of the microchamber array device. (A) Weighing the curing agent and prepolymer of PDMS. (B) Pouring the mixed and deaerated mixture into a tape-patterned Petri dish. There were some newly generated air bubbles in the polymer mixture. (C) Deaerating the mixture again in a vacuum chamber. The air bubbles were rising and burst on the top surface. (D) The deaerated and cured PDMS resin. (E) Peeling off the cured PDMS elastomer from the Petri dish. (F) Cutting off every piece of PDMS channel blocks using a flat-cable cutter. (G) Punching holes at each end of the PDMS channel using a biopsy punch. (H) The assembled device. The PDMS resin can adhere to the CYTOP surface. (I) Translucent microchamber array area inside the PDMS channel filled with the aqueous solution before chilling. (J) Transparent microchamber array area inside the PDMS channel after chilling. Please click here to view a larger version of this figure.
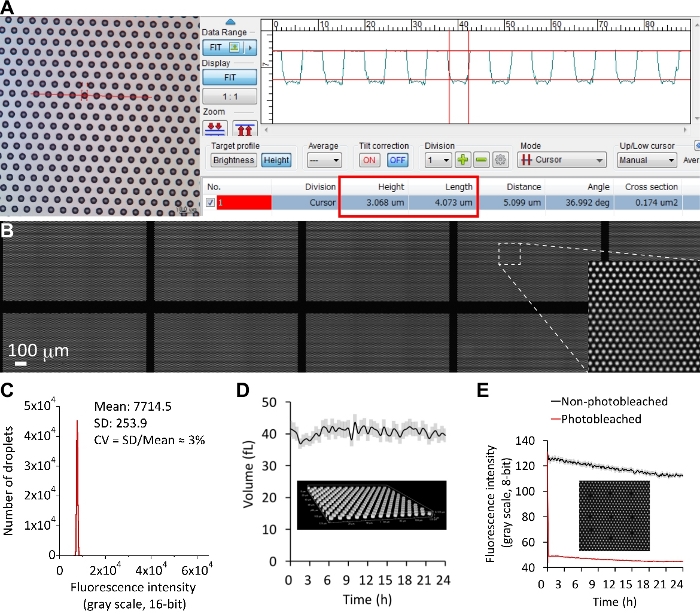
Figure 3: Ultra-uniform and ultra-stable femtoliter droplets. (A) 3D laser scanning confocal microscopy imaging for the fabricated substrate. The cylindrical microchambers in the example image showed a height of 3 μm and a diameter of 4 μm. (B) Uniform femtoliter droplets over a large area of the planar array. Only a partial area of the entire array was shown herein. A fluorescent solution (10 μM ATTO-514) was sealed in each droplet. (C) The size distribution of the droplets over an entire single array. The fluorescence intensity was used as an indicator of the droplet size. The CV was only 3%. (D) Volumetric measurement using confocal z-stack time-course data. The volume of droplets over the array was given by the microscope software (NIS-Elements, Nikon). (E) Fluorescence recovery after photobleaching. After the first frame, several droplets were completely photobleached using a confined laser beam of a confocal microscope. Their fluorescence intensity was recorded for 24 h, and no fluorescence recovery was observed (red line). The fluorescence intensity of other non-photobleached droplets in the same field of view was recorded in the black line. Error bars (translucent colors) were 1 SD for every time-point. Please click here to view a larger version of this figure.
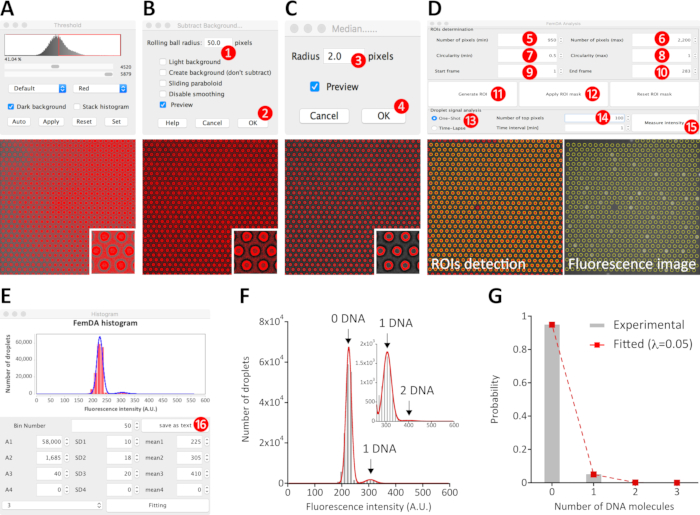
Figure 4: Analysis procedure of the end-point microscopic image data. The defocused bright-field image was used to extract the coordinate of every microchambers/droplets. Based on the intensity difference between the edge of microchambers (as foreground) and other areas (as background), the continuous and near-circular edge can be extracted from the background. Because of the uneven background distribution across the field of view (A), some image pre-processing is generally required to improve the quality of the binarizing output. After subtracting background (B), the background of every frame was uniformized. Empirically, the input value in the “Rolling ball radius” (step 1) was 20-50 for 2048 pixel × 2048 pixel images and 10-20 for 512 pixel × 512 pixel images. The larger the value, the shorter the processing time. To reduce the noise in the background-subtracted image, apply a median filter to the image (C). In general, the input value in the Radius (step 3) was 1-2 pixels. As shown in the magnified insert, the background-subtracted and filtered image can be nicely binarized using the build-in threshold plugin so that the foreground corresponding to the edges as well as the region of interests (ROIs) can be accurately recognized. The ROIs detection for all frames of the image was carried out by using the installed homemade plugin FemDA Analysis (D). The pixel size (steps 5 and 6) and circularity (steps 7 and 8) of ROIs, the frames that we want to put into the calculation (steps 9 and 10) were manually defined according to the actual data. After clicking Generate ROI (step 11) and waiting, the coordinate of every ROI across the input frames was determined. After clicking the Apply ROI mask (step 12), every detected ROI was enclosed and highlighted by a yellow line. After opening the fluorescence image and clicking the Apply ROI mask again, the determined ROIs mask was applied to the fluorescence image. The Number of top pixels (step 14) specified the number of top intensity-ranked pixels of each ROI for the calculation of the mean intensity of the respective droplets. After clicking Measure intensity (step 15) and waiting, the histogram was generated (E). The histogram can be fitted with a sum of Gaussian distributions using the parameters available in the histogram window. Alternatively, the mean intensity data can be exported to a text file (step 16) and analyzed by other software (F). (G) The probability of occurrence of droplets containing different numbers of DNA molecules in the given array. The histogram (grey color) was nicely fitted by a Poisson distribution (red dashed line) with an average of 0.05 DNA molecules per droplet, as expected for a random distribution of DNA molecules. Please click here to view a larger version of this figure.
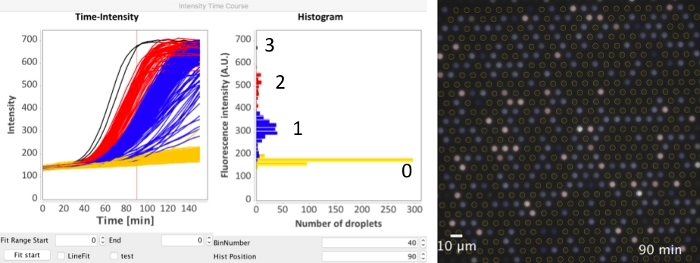
Figure 5: Analysis of the time-course microscopic image data. The detected ROIs (following the steps 1-12 of Figure 4) was directly applied to the time-course data. In step 13 of Figure 4, select Time-lapse and input the actual time-interval (in minutes) between adjacent frames in Time-interval [min]. After clicking Measure intensity (step 15 of Figure 4) and waiting, the time-intensity plot and the histogram (the same as Figure 4E) were generated. The Hist position specified the time-point of the histogram, which was shown as a vertical red line. The plugin also supports specifying colors for every trace line. Yellow: 0 DNA; blue was 1 DNA; red: 2 DNA; black: 3 DNA. The time-course data can also be exported to a text file. Please click here to view a larger version of this figure.
Supplementary Movie 1. Please click here to download this movie.
Discussion
The highly quantitative measurement based on the highly uniform, stable, and biocompatible droplets in FemDA enabled the discrete distribution, the unique feature of our study differing from others. We systematically optimized and detailed the microfabrication and droplet formation processes in this paper. There are several critical steps in the established protocol.
First, the uniform coating of highly viscous CYTOP polymer on the rectangular thin glass substrate largely determines the quality of the resulting substrate. Given the generally short working distance of the objective lens with high magnifications (see step 6.1), the thin cover glass must be used. A high-quality spin-coating on a thin and flexible substrate is generally tricky. We have not yet tested all possible designs but have found that the multi-hole vacuum chuck with the same rectangular dimension did work well for the spin-coating. It is important to drop the CYTOP polymer at the center of the cover glass substrate and immediately start the spin-coating (see step 1.3.2). The degree of the difficulty is less related to the shape of the substrate but the viscosity of the polymer. The CYTOP product line offers several different concentrations of the commercially available package. The accompanying diluent in the package can also be used to dilute the original product to a lower viscosity if needed. CYTOP 816 that we used in the experiment is the commercial product with the highest concentration capable of stably dissolving the polymer in the solvent. The thicker coating requires lower spin speed or multiple rounds of coating and curing. A plot of characteristic thickness vs. spin speed curve is highly recommended when using a new spin coater in a new environment.
Second, the appropriate humidity of the cleanroom is crucial for ideal photolithography as well as the complete removal of the photoresist at the specified position. The photochemical reaction in photolithography uses H2O as one of the reactants. However, the softbake at the temperature higher than the boiling temperature of H2O removes H2O content from the coated photoresist. The “dried” photoresist must take time to absorb H2O again from the air (see step 1.4.6). This point is often overlooked. Given that many laboratories may only have some simplified cleanroom facilities without humidity control, the humidity would fluctuate dramatically over the season or be affected by the weather. A low-cost solution is to use a home-use humidifier or dehumidifier in the cleanroom. The fully exposed CYTOP layer after development can be selectively and fully removed by RIE, fully exposing the glass bottom. Eventually, the hybrid structure of the hydrophilic glass bottom and the hydrophobic CYTOP sidewall is important for stably trapping aqueous solution to the femtoliter space.
Third, the newly found oils (ASAHIKLIN AE-3000, Fomblin Y25) and surfactant (SURFLON S-386) showed unprecedented sealing performance for the fluoropolymer reactor13. The injection of the flush oil must be carried out on the chilled flat metal block (see step 5.2); otherwise, droplets near the inlet of the channel cannot be formed. The injection of the second sealing oil can be carried out at RT (see step 5.5). These oils and surfactants have never been used in biological research before. No better alternatives have been found for now. To expand the arsenal of the useful combination of oils and surfactants for droplet preparation, the new member of the oils (or the similar ones in the same product line) and the surfactant (or the similar ones in the same product line) are worthy of trying to be applied in microfluidic droplet systems.
Here are two additional points to notice. One is the fact that there is a time lag among ROIs during the microscopy imaging. The total imaging time for one cycle is mainly dependent on the array size and the magnification of the objective lens. The exposure time, shutter speed, and the moving speed of the motorized stage are also minor factors. As a rule of thumb, imaging 106 droplets at 60× magnification would at least require 10-20 minutes. This inevitable time lag introduces some errors in data analysis, which should always be taken into careful consideration. The other one is the fact that the total number of droplets on a single substrate would not exceed 108–109, even increasing the density of the droplets or reducing the individual size of the droplet. This is because the size of the glass substrate that can be handled by the mask aligner (for 4-inch wafers) is limited. Further increasing the size of the cover glass is not recommendable because a large, thin, and fragile glass substrate is hard to handle.
FemDA can be considered as a super miniaturized microtiter plate. Any biochemical reactions that have been carried out in 96-well microtiter plates could be tried to conduct using FemDA. It can surely be used for enzyme activity measurement without complicated protein purification. It could also be compatible with digital polymerase chain reaction (digital PCR) and digital enzyme-linked immunosorbent assay (digital ELISA)31,49. The small volume, large array, and high stability would bring some advantages over existing digital bioassay methods. The FemDA system capable of resolving different numbers of template DNA molecules in femtoliter droplets has already shown the power on accurate protein screening13. FemDA is a new in vitro compartmentalization system capable of rapidly evolving a variety of enzymes. With the advances in bioinformatics and library construction techniques, the era of a prompt on-demand creation of high-performance proteins will be surely coming.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by JSPS KAKENHI grant number JP18K14260 and the budget of Japan Agency for Marine-Earth Science and Technology. We thank Shigeru Deguchi (JAMSTEC) and Tetsuro Ikuta (JAMSTEC) for providing the characterization facilities. We thank Ken Takai (JAMSTEC) for commercial software support. The microfabrication was conducted at Takeda Sentanchi Supercleanroom, The University of Tokyo, supported by "Nanotechnology Platform Program" of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, Grant Number JPMXP09F19UT0087.
Materials
| (3-aminopropyl)triethoxysilane | Sigma-Aldrich | 440140 | |
| 1 mL syringe | Terumo | SS-01T | |
| 2-propanol | Kanto Chemical | EL grade | EL: for electronic use. |
| 3D laser scanning confocal microscope | Lasertec | OPTELICS HYBRID | Other similar microscopes (e.g., Keyence VK-X1000, Olympus LEXT OLS5000) are also applicable. |
| 50 mL syringe | Terumo | SS-50LZ | |
| 6,8-difluoro-4-methylumbelliferyl phosphate | Thermo Fisher Scientific | D6567 | Prepare a 5 mM stock solution in dimethyl sulfoxide |
| Acetone | Kanto Chemical | EL grade | EL: for electronic use. Purity 99.8%. |
| Air blower | Hozan | Z-263 | |
| Aluminum block | BIO-BIK | AB-24M-02 | |
| Aluminum microtube stand | BIO-BIK | AB-136C | |
| ASAHIKLIN AE-3000 | AGC | (Test sample) | Free test sample may be available upon inquiry to AGC. |
| BEMCOT PS-2 wiper | Ozu | 028208 | |
| Biopsy punch with plunger | Kai | BPP-10F | |
| Cover glass | Matsunami Glass | No. 1 (24 mm × 32 mm, 0.13~0.17 mm thickness) | Size-customized. |
| Cover glass staining rack | Nakayama | 803-131-11 | |
| CRECIA TechnoWipe clean wiper | Nippon Paper Crecia | C100-M | |
| Cutting mat | GE Healthcare | WB100020 | |
| CYTOP | AGC | CTL-816AP | |
| Deaeration mixer | Thinky | AR-100 | |
| Desktop cutter | Roland | STIKA SV-8 | |
| Developer | AZ Electronic Materials | AZ 300 MIF | AZ Electronic Materials was now acquired by Merck. Other alkaline developers may be also applicable but should require optimization of development conditions (time, temperature, etc.) |
| Double-coated adhesive Kapton film tape | Teraoka Seisakusho | 7602 #25 | |
| Ethanol | Kanto Chemical | EL grade | EL: for electronic use. Purity 99.5%. |
| Fiji | Version: ImageJ 1.51n | ||
| Flat-cable cutter | Tokyo-IDEAL | MT-0100 | |
| Fomblin oil | Solvay | Y25, or Y25/6 | Free test sample may be available upon inquiry to Solvay. Fomblin Y25/6 is an alternative if Y25 is not readily available. |
| Hot plate | AS ONE | TH-900 | |
| Injection needle | Terumo | NN-2270C | 22G × 70 mm |
| Inverted fluorescence microscope | Nikon | Eclipse Ti-E | Epifluorescence specification, CCD or sCMOS camera, motorized stage, autofocus system, and high NA objective lens are required. |
| KaleidaGraph | Synergy | Version: 4.5 | |
| Mask aligner | SUSS | MA-6 | Other mask aligners are also applicable as long as the vacuum contact mode is avaliable. |
| MICROMAN pipette | GILSON | E M250E | Capillary piston tip: CP250 |
| Microsoft Excel | Microsoft | Version: 16.16.15 | |
| Mini vacuum chamber | AS ONE | MVP-100MV | |
| Nuclease-free water | NIPPON GENE | 316-90101 | |
| Parafilm | Amcor | PM-996 | |
| PCR tube | NIPPON Genetics | FG-021D/SP | |
| Petri dish | AS ONE | GD90-15 | Diameter 90 mm, height 15 mm. |
| Photoresist | AZ Electronic Materials | AZ P4903 | AZ Electronic Materials was now acquired by Merck. AZ P4620 is an alternative. |
| Plate reader | BioTek | POWERSCAN HT | |
| Polyethelene gloves | AS ONE | 6-896-02 | Trade name: Saniment. |
| PURExpress in vitro protein synthesis kit | New England Biolabs | E6800S or E6800L | For cell-free protein synthesis reaction. |
| Reactive-ion etching system | Samco | RIE-10NR | Other RIE systems are also applicable but should require optimization of RIE conditions (gas flow rate, chamber pressure, RF power, etching time, etc.) |
| RNase inhibitor | New England Biolabs | M0314S | |
| Scotch tape | 3M | 810-1-18D | |
| Sodium hydroxide solution | FUJIFILM Wako Pure Chemical | 194-09575 | 8 M concentration; danger. |
| Spin coater | Oshigane | SC-308 | |
| SURFLON S-386 surfactant | AGC | (Test sample) | Free test sample may be available upon inquiry to AGC. |
| SYLGARD 184 silicone elastomer | Dow | Sylgard184 | Chemical composition: polydimethylsiloxane. The default mixing ratio is base : curing agent = 10 : 1 (m/m). |
| Tweezers | Ideal-tek | 2WF.SA.1 2A |
|
| Ultrasonic cleaner | AS ONE | ASU-2M | |
| Vacuum chuck | Oshigane | (Customized) | Material: delrin; rectangular sample stage with multiple holes (48 holes, each with 1 mm diameter); the size is customzied to fit the size of the cover glass (24 mm × 32 mm). |
References
- Chiu, D. T., Lorenz, R. M., Jeffries, G. D. M. Droplets for ultrasmall-volume analysis. Analytical Chemistry. 81 (13), 5111-5118 (2009).
- Squires, T. M., Quake, S. R. Microfluidics: fluid physics at the nanoliter scale. Reviews of Modern Physics. 77 (3), 977-1026 (2005).
- Guo, M. T., Rotem, A., Heyman, J. A., Weitz, D. A. Droplet microfluidics for high-throughput biological assays. Lab on a Chip. 12 (12), 2146-2155 (2012).
- Zhu, P. A., Wang, L. Q. Passive and active droplet generation with microfluidics: a review. Lab on a Chip. 17 (1), 34-75 (2017).
- Griffiths, A. D., Tawfik, D. S. Miniaturising the laboratory in emulsion droplets. Trends in Biotechnology. 24 (9), 395-402 (2006).
- Tran, T. M., Lan, F., Thompson, C. S., Abate, A. R. From tubes to drops: droplet-based microfluidics for ultrahigh-throughput biology. Journal of Physics D: Applied Physics. 46 (11), 114004 (2013).
- Zhang, Y., Jiang, X., Fan, C. Microfluidic tools for DNA analysis. DNA Nanotechnology. , 113-153 (2013).
- Dubuc, E., et al. Cell-free microcompartmentalised transcription-translation for the prototyping of synthetic communication networks. Current Opinion in Biotechnology. 58, 72-80 (2019).
- Damiati, S., Mhanna, R., Kodzius, R., Ehmoser, E. K. Cell-free approaches in synthetic biology utilizing microfluidics. Genes. 9 (3), (2018).
- Lee, K. H., Kim, D. M. Applications of cell-free protein synthesis in synthetic biology: Interfacing bio-machinery with synthetic environments. Biotechnology Journal. 8 (11), 1292-1300 (2013).
- Supramaniam, P., Ces, O., Salehi-Reyhani, A. Microfluidics for artificial life: techniques for bottom-up synthetic biology. Micromachines. 10 (5), (2019).
- Bowman, E. K., Alper, H. S. Microdroplet-assisted screening of biomolecule production for metabolic engineering applications. Trends in Biotechnology. , (2019).
- Zhang, Y., et al. Accurate high-throughput screening based on digital protein synthesis in a massively parallel femtoliter droplet array. Science Advances. 5 (8), 8185 (2019).
- Silverman, A. D., Karim, A. S., Jewett, M. C. Cell-free gene expression: an expanded repertoire of applications. Nature Reviews Genetics. , (2019).
- Sakane, Y., Suzuki, Y., Kasagi, N. The development of a high-performance perfluorinated polymer electret and its application to micro power generation. Journal of Micromechanics and Microengineering. 18 (10), 104011 (2008).
- Sakakihara, S., Araki, S., Iino, R., Noji, H. A single-molecule enzymatic assay in a directly accessible femtoliter droplet array. Lab on a Chip. 10 (24), 3355-3362 (2010).
- Watanabe, R., et al. Arrayed lipid bilayer chambers allow single-molecule analysis of membrane transporter activity. Nature Communications. 5, 4519 (2014).
- Chiu, C., Lisicka-Skrzek, E., Tait, R. N., Berini, P. Fabrication of surface plasmon waveguides and devices in Cytop with integrated microfluidic channels. Journal of Vacuum Science & Technology B. 28 (4), 729-735 (2010).
- Krupin, O., Asiri, H., Wang, C., Tait, R. N., Berini, P. Biosensing using straight long-range surface plasmon waveguides. Optics Express. 21 (1), 698-709 (2013).
- Hanada, Y., Ogawa, T., Koike, K., Sugioka, K. Making the invisible visible: a microfluidic chip using a low refractive index polymer. Lab on a Chip. 16 (13), 2481-2486 (2016).
- Berry, S., Kedzierski, J., Abedian, B. Low voltage electrowetting using thin fluoroploymer films. Journal of Colloid and Interface Science. 303 (2), 517-524 (2006).
- Lin, Y. Y., et al. Low voltage electrowetting-on-dielectric platform using multi-layer insulators. Sensors and Actuators B-Chemical. 150 (1), 465-470 (2010).
- Kimura, H., Yamamoto, T., Sakai, H., Sakai, Y., Fujii, T. An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab on a Chip. 8 (5), 741-746 (2008).
- Yang, T. J., Choo, J., Stavrakis, S., de Mello, A. Fluoropolymer-coated PDMS microfluidic devices for application in organic synthesis. Chemistry-a European Journal. 24 (46), 12078-12083 (2018).
- de Gennes, P. G. Wetting: statics and dynamics. Reviews of Modern Physics. 57 (3), 827-863 (1985).
- Holtze, C., et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab on a Chip. 8 (10), 1632-1639 (2008).
- Wagner, O., et al. Biocompatible fluorinated polyglycerols for droplet microfluidics as an alternative to PEG-based copolymer surfactants. Lab on a Chip. 16 (1), 65-69 (2016).
- Mashaghi, S., Abbaspourrad, A., Weitz, D. A., van Oijen, A. M. Droplet microfluidics: a tool for biology, chemistry and nanotechnology. TrAC Trends in Analytical Chemistry. 82, 118-125 (2016).
- Mazutis, L., et al. Droplet-based microfluidic systems for high-throughput single DNA molecule isothermal amplification and analysis. Analytical Chemistry. 81 (12), 4813-4821 (2009).
- Galinis, R., et al. DNA nanoparticles for improved protein synthesis in vitro. Angewandte Chemie-International Edition. 55 (9), 3120-3123 (2016).
- Zhang, Y., Noji, H. Digital bioassays: theory, applications, and perspectives. Analytical Chemistry. 89 (1), 92-101 (2017).
- Mazutis, L., et al. Single-cell analysis and sorting using droplet-based microfluidics. Nature Protocols. 8 (5), 870-891 (2013).
- Courtois, F., et al. An integrated device for monitoring time-dependent in vitro expression from single genes in picolitre droplets. ChemBioChem. 9 (3), 439-446 (2008).
- Baret, J. C. Surfactants in droplet-based microfluidics. Lab on a Chip. 12 (3), 422-433 (2012).
- Agresti, J. J., et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proceedings of the National Academy of Sciences of the United States of America. 107 (9), 4004-4009 (2010).
- Fallah-Araghi, A., Baret, J. C., Ryckelynck, M., Griffiths, A. D. A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution. Lab on a Chip. 12 (5), 882-891 (2012).
- Duncombe, T. A., Dittrich, P. S. Droplet barcoding: tracking mobile micro-reactors for high-throughput biology. Current Opinion in Biotechnology. 60, 205-212 (2019).
- Qin, D., Xia, Y. N., Whitesides, G. M. Soft lithography for micro- and nanoscale patterning. Nature Protocols. 5 (3), 491-502 (2010).
- Mukhopadhyay, R. When PDMS isn’t the best. Analytical Chemistry. 79 (9), 3248-3253 (2007).
- Shaner, N. C., et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nature Methods. 10 (5), 407-409 (2013).
- Bradshaw, R. A., et al. Amino acid sequence of Escherichia coli alkaline phosphatase. Proceedings of the National Academy of Sciences of the United States of America. 78 (6), 3473-3477 (1981).
- Shimizu, Y., et al. Cell-free translation reconstituted with purified components. Nature Biotechnology. 19 (8), 751-755 (2001).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Mantilla, C. B., Prakash, Y. S., Sieck, G. C., Conn, P. M. Volume measurements in confocal microscopy. Techniques in Confocal Microscopy. , 143-162 (2010).
- Noji, H. Single-molecule counting of biomolecules with femtoliter dropret chamber array. The 17th International Conference on Solid-State Sensors, Actuators and Microsystems (TRANSDUCERS & EUROSENSORS XXVII. , 630-632 (2013).
- Zhang, Y., et al. Matrix-localization for fast analysis of arrayed microfluidic immunoassays. Analytical Methods. 4 (10), 3466-3470 (2012).
- Zhang, Y., et al. Two dimensional barcode-inspired automatic analysis for arrayed microfluidic immunoassays. Biomicrofluidics. 7 (3), 034110 (2013).
- Gonzalez, R. C., Woods, R. E. . Digital image processing. , (2018).
- Cohen, L., Walt, D. R. Single-molecule arrays for protein and nucleic acid analysis. Annual Review of Analytical Chemistry. 10 (1), 345-363 (2017).

