使用自适应氯化钙程序对大肠杆菌细胞的转化
English
Share
Overview
资料来源:纳塔利娅·马丁1号,安德鲁·范·阿尔斯特1号,瑞安农·勒维克1号,维克多·迪丽塔1号
1 密歇根州立大学微生物学和分子遗传学系
细菌有能力在称为水平基因转移的过程中交换遗传物质(脱氧核糖核酸,DNA)。结合外源性DNA提供了一种机制,细菌可以通过它获得新的遗传特性,使他们能够适应不断变化的环境条件,如抗生素或抗体的存在(1)或分子在自然栖息地发现(2) 水平基因转移有三种机制:转化、转导和共聚(3)。在这里,我们将专注于转化,细菌从环境中获取自由DNA的能力。在实验室中,转化过程有四个一般步骤:1) 制备合格细胞,2) 用DNA孵育合格细胞,3) 细胞恢复,4) 细胞的电镀,用于转化剂的生长(图1)。
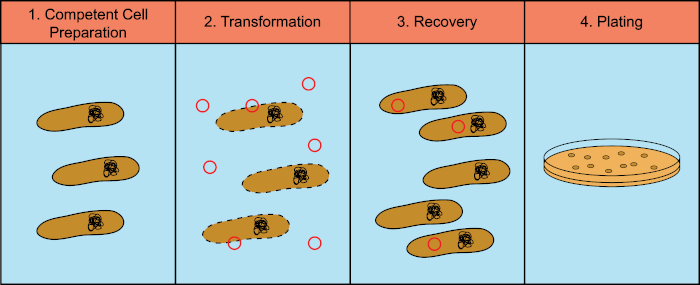
图 1:转换过程的一般步骤。转化过程有四个一般步骤:1) 制备合格细胞,2) 用DNA孵育,3) 细胞的恢复和 4) 电镀细胞的生长转化剂。
要发生转化,受体细菌必须处于称为能力的状态。有些细菌能够因某些环境条件而变得自然而然。然而,许多其他细菌不能自然地胜任,或者这个过程的条件还不得而知。将DNA引入细菌的能力具有一系列研究应用:生成感兴趣的DNA分子的多个副本,表达大量蛋白质,作为克隆过程中的组成部分,以及其他。由于转化对分子生物学的价值,有几个协议旨在使细胞在自然能力条件未知时人为地具有能力。两种主要方法用于制备人工能力细胞:1)通过细胞的化学处理,2)将细胞暴露于电脉冲(电穿孔)。前者使用不同的化学物质,具体取决于在DNA和细胞表面之间产生吸引力的程序,而后者使用电场在细菌细胞膜中产生孔隙,DNA分子可以通过这些孔进入。化学能力最有效的方法是用二价阳离子孵育,最显著的是钙(Ca2+)(4,5) 钙诱导能力是这里描述的程序 (6).该方法主要用于革兰氏阴性细菌的转化,是本方案的重点。
化学转化过程涉及一系列步骤,其中细胞暴露于阳离子诱导化学能力。这些步骤随后是温度变化 – 热休克 – 有利于由主管细胞(7)摄取外来DNA。细菌细胞包络呈负电荷。在革兰氏阴性细菌,如大肠杆菌,外膜是负电荷由于存在脂多糖(LPS)(8 )。这导致同样带负电荷的DNA分子排斥。在化学能力诱导中,带正电荷的钙离子中和这种电荷排斥,使DNA吸收到细胞表面(9)。钙处理和脱氧核糖核酸的孵育在冰上进行。随后,在较高温度(42°C)下进行孵育,进行热冲击。这种温度不平衡进一步有利于DNA的摄取。细菌细胞需要在中指数生长阶段,以承受热冲击处理;在其他生长阶段,细菌细胞对热量过于敏感,导致生存能力丧失,从而大大降低转化效率。
不同的DNA来源可用于转化。通常,在大肠杆菌的大多数实验室程序中,质粒、小圆形、双链DNA分子用于转化。要在转化后在细菌细胞中维持质粒,它们需要包含复制的来源。这使得它们可以在细菌细胞中独立于细菌染色体复制。并非所有的细菌细胞在转化过程中都得到转化。因此,转化产生转化细胞和非转化细胞的混合物。为了区分这两个群体,使用一种选择方法来识别获得质粒的细胞。疟原虫通常含有可选择的标记物,这些标记是编码一种具有生长优势的特征的基因(即对抗生素或化学物的抗药性或从生长辅助体中拯救)。转化后,细菌细胞被镀在选择性培养物上,这只允许转化细胞的生长。如果细胞转化,质粒对给定抗生素具有抗药性,选择性培养基将是含有该抗生素的生长介质。有几种不同的方法可以用来确认在选择性培养基中生长的菌落是转化剂(即已经合并了质粒)。例如,质粒可以使用质粒制备方法(10)从这些细胞中恢复,并消化以确认质粒大小。或者,菌落PCR可用于确认存在感兴趣的质粒(11)。
本实验的目的是利用氯化钙程序(12)的适应,制备大肠杆菌DH5+化学能力细胞,并用质粒pUC19对其进行转化,以确定转化效率。大肠杆菌菌株DH5+是分子生物学应用中常用的菌株。由于其基因型,特别是recA1和endA1,这种菌株可以提高插入稳定性,并在随后的制剂中提高质粒DNA的质量。由于转化效率随着DNA尺寸的增加而降低,因此,由于质粒pUC19体积小(2686 bp),因此在本协议中使用了质粒pUC19(参见https://www.mobitec.com/cms/products/bio/04_vector_sys/standard_cloning_vectors.html矢量映射)。pUC19对青霉素具有抗药性,因此,这是用于选择的抗生素。
Procedure
Results
Although TE is dependent on many factors, non-commercial competent cell preparations, like this one, normally yield 106 to 107 transformants per microgram of plasmid. Therefore, this preparation, with a TE = 2.46 x 108 cfu/µg, yielded a TE well beyond the expected range. Additional protocols are available for making supercompetent cells when higher transformation efficiencies are required for a given application (13).
Analysis of the digestion of the plasmid DNA recovered from the transformed cells indicated that this plasmid has the expected size of pUC19 DNA (2686 bp).
Applications and Summary
Transformation is a powerful method for introducing exogenous DNA into bacterial cells that is key to many molecular biology applications in the laboratory. Additionally, it plays a major role in nature by allowing bacterial cells to exchange genetic material that could result in increased genetic variation and allow for acquisition of different beneficial traits for survival under a wide range of conditions. Many bacterial strains encode the genes required for natural competence. However, the conditions in which these genes are induced are still unknown. Further research is required to determine these conditions.
References
- Croucher, N. J. et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 331 (6016):430-434. (2011)
- Borgeaud, S. et al. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science. 347(6217):63-67. (2015)
- Burmeister, A. R. Horizontal Gene Transfer. Evol Med Public Health. 2015 (1):193-194. (2015)
- Weston A, Brown MG, Perkins HR, Saunders JR, Humphreys GO. Transformation of Escherichia coli with plasmid deoxyribonucleic acid: calcium-induced binding of deoxyribonucleic acid to whole cells and to isolated membrane fractions. J Bacteriol. 145 (2):780-7. (1981)
- Dagert M, Ehrlich SD. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 6 (1):23-8. (1979)
- Asif A, Mohsin H, Tanvir R, and Rehman Y. Revisiting the Mechanisms Involved in Calcium Chloride Induced Bacterial Transformation. Front Microbiol. 8:2169. (2017)
- Panja S, Aich P, Jana B, Basu T. How does plasmid DNA penetrate cell membranes in artificial transformation process of Escherichia coli? Mol Membr Biol. 25 (5):411-22. (2008)
- Silhavy, TJ, Kahne D, Walker S. The Bacterial Cell Envelope. Cold Spring Harb Perspect Biol. 2 (5): a000414. (2010)
- Panja S, Aich P, Jana B, Basu T. (2008) Plasmid DNA binds to the core oligosaccharide domain of LPS molecules of E. coli cell surface in the CaCl2-mediated transformation process. Biomacromolecules. 9 (9):2501-9.
- JoVE Science Education Database. Basic Methods in Cellular and Molecular Biology. Plasmid Purification. JoVE, Cambridge, MA. (2018)
- Bergkessel M and Guthrie C. Colony PCR. Methods in Enzymology. 529: 299-309. (2013)
- Sambrook J and Russell DW. Molecular Cloning A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.Protocol 25 (1.116-118). (2001)
- Wirth R, Friesenegger A, Fiedler S. Transformation of various species of gram-negative bacteria belonging to 11 different genera by electroporation. Molecular & General Genetics. 216 (1): 175-7. (1989)
Transcript
Bacteria are remarkably adaptable and one mechanism which facilitates this adaptation is their ability to take in external DNA molecules. One type of DNA that bacteria can uptake is called a plasmid, a circular piece of DNA that frequently contains useful information, such as antibiotic resistance genes. The process of bacteria being modified by new genetic information incorporated from an external source is referred to as transformation. Transformation can easily be performed in the laboratory using Escherichia coli, or E. coli.
In order to be transformed, E. coli cells must first be made competent, which means capable of taking in DNA molecules from their environment. The protocol for accomplishing this is surprisingly simple, a short incubation of the cells in a calcium chloride solution. This incubation causes the cells to become permeable to DNA molecules. After the cells are pelleted by centrifugation, the supernatant is removed. The plasmid DNA is now added to the competent cells. After incubating the cells with DNA, the mix is briefly heated to 42 degrees Celsius, followed by rapid cooling on ice. This heat shock causes the DNA to be transferred across the cell’s wall and membranes. The cells are then incubated in fresh media. Then, the bacteria are placed at 37 degrees to allow them to reseal their membranes and express resistant proteins.
Those cells which have taken in the plasmids will faithfully copy the DNA and pass it to their progeny and express any proteins that might be encoded by it, including antibiotic resistance mediators. Those resistance genes can be used as selectable markers to identify bacteria which have been successfully transformed because cells that have not taken up the plasmid will not express the resistance gene product. This means that when the cells are plated on a solid medium which contains the appropriate antibiotic, only cells that have taken up the plasmid will grow. Transformation of the cells in a growing colony can be further confirmed by culturing those cells in liquid media overnight to increase the yield before extracting the DNA from the sample. Once the DNA is isolated, a diagnostic restriction enzyme digest can be carried out. Because restriction enzymes cut DNA in predictable locations, running these digests on a gel should show a predictable pattern if the desired plasmid was successfully transformed. For example, if pUC19 is prepared and cut with the restriction enzyme HindIII, a single band of 2686 nucleotides should be seen on the gel.
In this lab, you will transform E. coli strain DH-5 Alpha with pUC19, and then confirm the successful transformation by DNA gel electrophoresis.
Before starting the procedure, put on the appropriate personal protective equipment, including a lab coat and gloves. Next, sterilize the workspace with 70% ethanol.
Now, prepare chemically competent cells by depositing a loopfull of bacteria onto a sterile LB agar plate and streaking the bacteria with a new loop. Then, incubate the plate at 37 degrees Celsius overnight. The next day, sterilize the bench top with 70% ethanol again, and remove the plate from the incubator.
Inoculate a single, well-isolated colony into 3 milliliters of LB broth in a tube with a sterile loop. Then, grow the culture at 37 degrees Celsius overnight, with shaking at 210 RPM. The next day, measure the optical density of the overnight culture with a spectrophotometer. Then, add 100 milliliters of LB broth to a one-liter flask, and inoculate it with the overnight culture at an optical density of 0. 01. Now, incubate the culture at 37 degrees Celsius with shaking, and check the OD600 every 15 to 20 minutes until the culture reaches mid-exponential growth phase.
After approximately three hours, transfer 50 milliliters of the culture to two ice-cold polypropylene bottles. Then, place the bottles back on ice for 20 minutes to cool. Next, recover the cells via centrifugation. Discard the supernatants and place the bottles upside down on a paper towel. Next, resuspend the bacterial pellet in five milliliters of ice-cold calcium chloride magnesium chloride solution and swirl carefully until the pellet has dissolved completely. Then, add another 25 milliliters of the solution to the dissolved bacterial pellet. Resuspend the other bacterial pellet as previously demonstrated. After this, repeat the centrifugation, and remove the supernatants.
If the competent cells are going to be directly transformed, resuspend each bacterial pellet in two milliliters of an ice-cold 0.1 molar calcium chloride solution by swirling the tubes carefully. To begin the transformation procedure, transfer 50 microliters of competent cells to two labeled 1.5 milliliter polypropylene tubes. Then, add one microliter of pUC19 plasmid DNA to one of the tubes. Mix gently, avoiding bubble formation, and incubate both tubes for 30 minutes on ice. After incubation, transfer the tubes to a heat block and incubate at 42 degrees Celsius for 45 seconds. Immediately transfer the tubes to ice, and incubate for two minutes. Now, add 950 microliters of SOC media to each tube and incubate them for one hour at 37 degrees Celsius to allow the bacteria to recover, and express the antibiotic resistant marker encoded in the plasmid.
To make a 1 to 100 dilution, add 990 microliters of SOC media and 10 microliters of cell suspension to a 1.5 milliliter tube. Then, make a 1 to 10 dilution by adding 900 microliters of SOC media and 100 microliters of cell suspension to a 1.5 milliliter tube. Next, plate 100 microliters of the diluted cell suspensions and 100 microliters of the negative control, onto separate selective plates containing ampicillin using a spreader and incubate the plates at 37 degrees Celsius for 12 to 16 hours. After incubation, count the colony-forming units, or CFUs, per plate, obtained through transformation, and record these data. To verify that the transformants have the pUC19 plasmid, pick a single, well-isolated colony from a plate with a sterile loop, and introduce it to a tube containing 3 milliliters of LB broth. Then, incubate the culture at 37 degrees Celsius with shaking, overnight. The next day, use a DNA mini prep kit to isolate DNA from 3 milliliters of the culture, according to the manufacturer’s instructions. After completing the DNA mini prep, digest the 1 microgram of purified pUC19 with a restriction enzyme at 37 degrees Celsius for 1 hour. Now, load 20 microliters of a molecular weight ladder, 1 microgram of digested plasmid DNA, and 1 microgram of undigested plasmid DNA into consecutive wells of a 1% agarose gel containing 1 microgram per milliliter ethidium bromide. Then, run the gel for 1 hour at 95 volts. Finally, visualize the gel with a UV illuminator.
In this experiment, E. coli DH5 Alpha chemically competent cells were prepared using an adaptation of the calcium chloride procedure, and then transformed with the plasmid pUC19 to determine transformation efficiency. To calculate the transformation efficiency, use the recorded CFU counts for the 1 in 100 and 1 in 10 dilutions, and any other dilutions with CFU counts between 30 and 300. First, the recorded CFU count, 246 in this example, is divided by the amount of DNA, .0001 micrograms here, that was plated. Then, this number is divided by the dilution factor used to give the transformation efficiency in CFUs per microgram. In this example, a 1 to 10 dilution was used and 100 microliters of a 1 milliliter solution was plated, giving a final dilution factor of 0.01. In the undigested plasmid lane, the circular DNA may appear as two or three different bands of varying brightness. This is because the circular, uncut DNA may exist in several different conformation states, such as supercoiled, open circle, or more linear, and each of these move through the gel at different rates. Analysis of the recovered plasmid DNA digestion indicated that the plasmid used has an expected size of pUC19 DNA, 2,686 base pairs.
