結合:アンピシリン耐性をドナーからレシピエント大腸菌に移す方法
English
Share
Overview
ソース: アレクサンダー S. ゴールド1, トーニャ M. コルピッツ1
1ボストン大学医学部微生物学科、国立新興感染症研究所、ボストン、マサチューセッツ州
1946年にレダーバーグとテイタムによって最初に発見された共役は、2つの細菌細胞間の直接的な物理的接触に依存する細菌間の水平遺伝子伝達の一形態である(1)。形質転換やトランスダクションなどの他の形態の遺伝子導入とは異なり、結合はDNAがドナー細胞からレシピエント細胞に一方向的に分泌される自然発生プロセスです。この方向性と細菌の遺伝的多様性を高めるこのプロセスの能力は、抗生物質耐性の最近の上昇に大きく貢献していると考えられている細菌「交配」の一形態としての評判を共役に与えました。バクテリア(2、3)。選択的圧力(例えば抗生物質の使用)を用いることで、実験室での使用のためにコンジュゲーションが操作され、細菌間の水平遺伝子移動、場合によっては細菌から酵母、植物、動物への強力なツールとなっています。セル (4)。実験室での応用とは別に、結合による細菌・ユーカリオテ遺伝子の伝達は、多くの可能なバイオテクノロジーアプリケーションと自然に起こる影響を持つDNA伝達のエキサイティングな手段です(5)。
コンジュゲーションは「2段階機構」(6)によって働くと考えられている。まず、DNAを移送する前に、ドナー細胞はレシピエントと直接細胞間接触を行わなければならない。このプロセスは、グラム陰性細菌で最もよく特徴付けられており、その中で最も研究されているのは大腸菌である。細胞間接触は、性ピラスとして知られているドナー上の細胞外フィラメントの複雑なネットワークの存在によって確立され、F(不妊)因子(7,8)として知られる転写可能な遺伝子によってコードされる共役元素である。ドナーとレシピエントとの接触を確立することに加えて、いくつかのタンパク質は、レシピエント細胞質に性ピラスを介して輸送され、2つの細胞間のIV分泌系(T4SS)導管を形成し、第2工程に必要な構造である。DNA伝達(6)。この性ピルスの機能とDNAの転がり円複製を組み合わせることにより、ドナー細胞は、プラスミドまたはトランスポゾンなどのトランスポーザブル要素の形でDNAを「シュートアンドポンプ」モデル(6)によってレシピエントに移すことができる。この場合、「シューティング」は、パイロットタンパク質の輸送であり、連結DNAを用いて、T4SSによってレシピエント細胞に送られ、「ポンピング」はレシピエントへのDNAの活性輸送であり、T4SSに依存し、結合タンパク質によって触媒されるプロセスである(6)。このプロセスで使用される機械は、リラクサーゼ、合体対形成複合体、およびIV型結合タンパク質をコードするシスおよびトランス遺伝子のDNAによって提供されなければならない転移配列(oriT)の起源から構成され、シスまたはトランス(9)に存在することができます。このリラクサーゼは、oriT配列内のnic部位を切断し、転移した鎖の5’末端に生有して、他の補助タンパク質との一本鎖DNAリラクサーゼ複合体であるリラクソームを産生する(9)。いったん形成されると、リラクソームは交配対形成複合体に接続し、IV型結合タンパク質を介して、SsDNA-relaxase複合体をT4SS(10)によってレシピエント細胞に移植することを可能にする。レシピエントの細胞質に入ると、DNAはレシピエントゲノムに統合することも、プラスミドの形で別々に存在することも、その遺伝子の発現を可能にする。
この実験では、広く使用されている共役ドナー株大腸菌WM3064を、レシピエント株大腸菌J53に対するアンピシリン耐性をコードする遺伝子コードを伝達するために用いられた。グラム陰性菌の両方の株はテトラサイクリンに耐性であったが、ドナー株WM3064のみがアンピシリン耐性の遺伝子を持ち、pWD2-oriTシャトルベクターでコードされ、ジアミノピメル酸(DAP)に対して下方栄養性であった(11-13)。この実験は、ドナーおよびレシピエント株の調製、続いてコンジュゲーションによるドナーからレシピエントへのアンピシリン耐性遺伝子の移植の2つの主要なステップからなる(図1)。
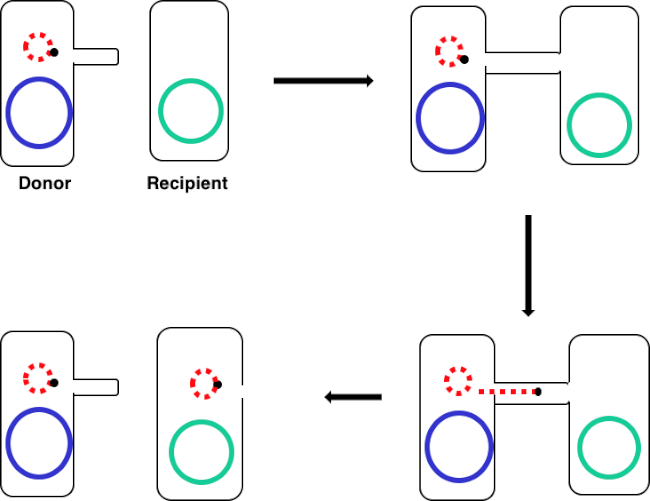
図 1: コンジュゲーション回路図。この回路図は、プラスミドの正常な転移を示し、転移可能なDNA要素の一例に過ぎず、共役を用いてドナー細胞からレシピエント細胞へ送り込む。性ピラスを介してドナー細胞によってレシピエント細胞と接触すると、プラスミドは転がり円複製によって複製し、2つの細胞を結合する多タンパク質複合体を通って移動し、レシピエント細胞に新しい全長プラスミドを形成する。
ドナー細胞とレシピエント細胞の混合物をインキュベートすることにより、テトラサイクリンおよびDAPの存在下でこれらの細胞を連続的にめっきし、これはアンピシリン耐性遺伝子の正常な転写を可能にした。テトラサイクリンとアンピシリンの存在下でこの混合物から増殖した細胞を連続的にめっきし、DAPの欠如とアンピシリン耐性遺伝子を得ていない可能性のある任意のレシピエント細胞の欠如のためにすべてのドナー細胞を除去し、厳密にレシピエントJ53株を得たアンピシリン耐性を獲得した細菌(図2)。一旦行われると、アンピシリン耐性遺伝子の正常な転写がPCRによって確認された。大腸菌のJ53株はpWD2-oriTを含み、アンピシリンに対して耐性があり、この耐性をコードする遺伝子はPCRによって検出可能であった。しかし、もし失敗した場合、アンピシリン耐性遺伝子の検出はなく、アンピシリンはJ53株に対する有効な抗生物質として依然として機能するだろう。
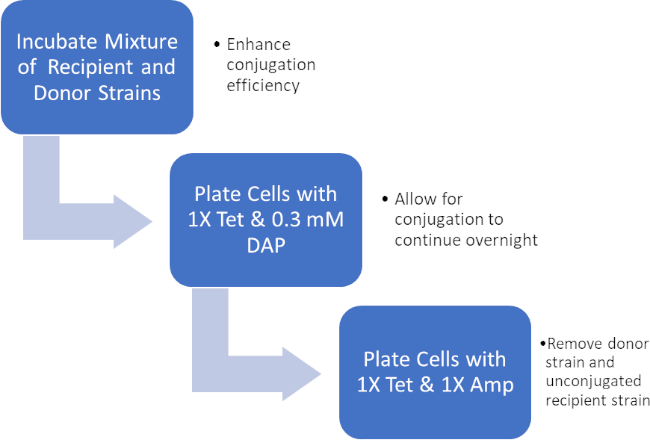
図 2: プロトコルの回路図。この回路図は、提示されたプロトコルの概要を示しています。
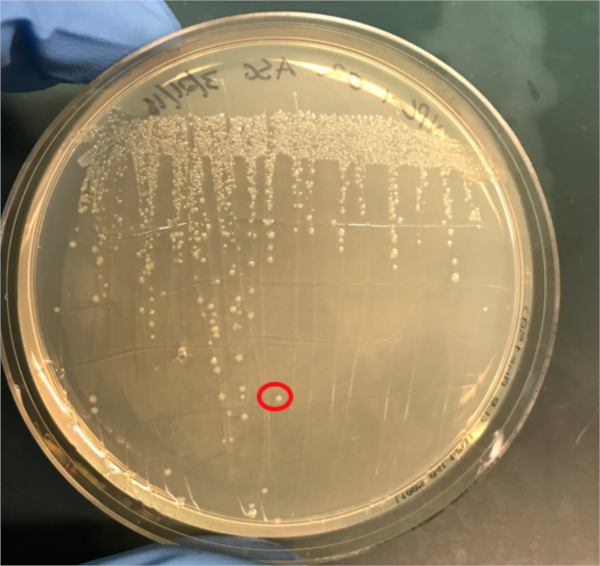
図3A:PCRによるコンジュゲーションの成功の確認A)共役および陰性対照試料の冷凍庫ストックを寒天プレート上にストリークアウトし、DNA分離のためにコロニー(赤色)を選択した。
Procedure
Results
If conjugation was successful, a 500 base-pair sized band PCR product will be observed in the well in which PCR reaction 1 was loaded (Well #2 in Figure 3B), while no bands will be observed in the well in which PCR reaction 3 was loaded (Well #4 in Figure 3B). The presence of this band confirms the successful transfer of the ampicillin resistance gene, thereby conferring ampicillin resistance to the J53 strain of E. coli.
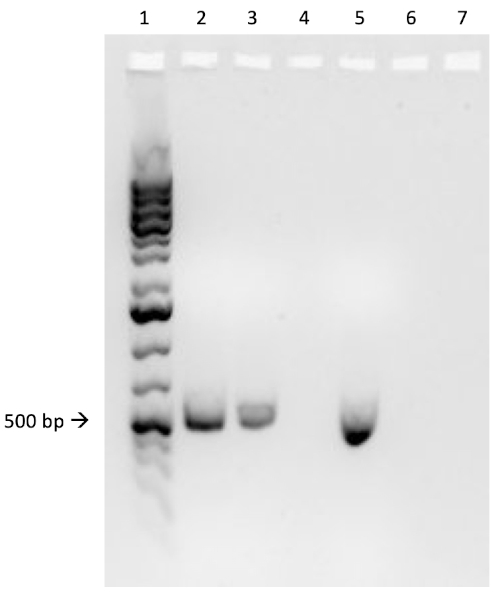
Figure 3B: The confirmation of successful conjugation by PCR. B) PCR analysis was done using DNA isolated from the select colony. The contents of each well are as follows: 1) DNA ladder, 2) Conjugation DNA and ampicillin primers, 3) Conjugation DNA and housekeeping primers, 4) Negative control DNA and ampicillin primers, 5) Negative control DNA and housekeeping primers, 6) No DNA and ampicillin primers, and 7) No DNA and negative control primers. The presence of a ~ 500 base-pair band PCR product from PCR reaction 1 (well 2), and the lack of this product from PCR reaction 3 (well 4), confirms successful conjugation.
Applications and Summary
Conjugation is a naturally occurring process of horizontal gene transfer that relies on the direct cell-to-cell contact of a donor cell and a recipient cell. This process is shared among all kinds of bacteria and has been instrumental in bacterial evolution, most notably antibiotic resistance. In the lab, conjugation can be used as an effective method of gene transfer that is much less disruptive when compared to other techniques. Outside of the laboratory, the ability to transfer DNA from bacteria to eukaryotes via conjugation offers an exciting new avenue of gene therapy and understanding the implications of these naturally occurring gene transfers, for example the relationship between bacterial infection and cancer, is a rapidly emerging area of research.
References
- Lederberg J, Tatum, E.L. Gene recombination in Escherichia coli Nature. 1946;158:558.
- Holmes R.K. J, M.G. Genetics: Exchange of Genetic Information. 4th Edition ed. Baron S, editor. Galveston, TX: University of Texas Medical Branch at Galveston; 1996.
- Cruz F, Davies, J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends in Microbiology. 2000;8:128-33.
- Llosa M, Cruz, F. Bacterial conjugation: a potential tool for genomic engineering. Ressearch in Microbiology. 2005;156:1-6.
- Lacroix B, Citovsky, V. Transfer of DNA from Bacteria to Eukaryotes. mBio. 2016;7(4):1-9.
- Llosa M, et al. Bacterial conjugation: a two-step mechanism for
- DNA transport. Molecular Microbiology. 2002;45:1-8.
- Grohmann E, Muth, G., Espinosa, M. Conjugative Plasmid Transfer in Gram-Positive Bacteria. Microbiology and Molecular Biology Reviews. 2003;67:277-301.
- Firth N, Ippen-Ihler, K, Skurray, RA. Structure and function of the F factor and mechanism of conjugation. Escherichia coli and salmonella: cellular and molecular biology. 1996;2:2377-401.
- Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EPC, De La Cruz F. Mobility of Plasmids. Microbiology and Molecular Biology Reviews. 2010;74(3):434-52.
- Cascales E. Definition of a Bacterial Type IV Secretion Pathway for a DNA Substrate. 2004;304(5674):1170-3.
- Wang P, Yu Z, Li B, Cai X, Zeng Z, Chen X, et al. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Microbial Cell Factories. 2015;14(1):11.
- Yi H, Cho YJ, Yong D, Chun J. Genome Sequence of Escherichia coli J53, a Reference Strain for Genetic Studies. Journal of Bacteriology. 2012;194(14):3742-3.
- Baumann RLB, E. H.; Wiseman, J. S.; Vaal, M.; Nichols, J. S. Inhibition of Escherichia coli Growth and Diaminopimelic Acid Epimerase by 3-Chlorodiaminopimelic Acid. Antimicrobial Agents and Chemotherapy 1988;32:1119-23.
- Rocha D, Santos, CS, Pacheco LG. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek. 2015;108:685-93.
Transcript
Bacterial cells, such as E. coli, are able to transfer genetic information from cell-to-cell. Conjugation differs from other mechanisms of DNA transfer, such as transduction or transformation, in that it requires physical contact between the cells.
To proceed, conjugation requires a donor cell that expresses the fertility, or F, factor and a recipient cell without it, an F minus cell. The process requires two steps. The first is the establishment of direct cell-to-cell contact. To do this, the donor cell generates an extracellular filamentous structure called a sex pilus. It is named this since conjugation is a form of mating for asexually reproducing bacteria, but it should be noted that it is not true sexual reproduction as no gametes are exchanged and no offspring are formed.
The second step is delivery of DNA to the recipient cell. After the sex pilus establishes contact between two cells, a conduit called the Type IV secretion system is built allowing for the transfer of DNA. The donor cell then begins to replicate the extrachromosomal DNA that will be transferred selected based on the presence of a genetic element known as the OriT or origin of transfer. One end of the newly replicated DNA is threaded into the conduit through DNA protein binding. As the DNA is further replicated, it is pumped through the channel, facilitated by a complex of proteins encoded by genes located close to the OriT. Once the DNA is fully transferred, it will either form an extra chromosomal plasmid, or it may integrate into the chromosome of the recipient cell. Whichever the endpoint of the transferred DNA, the genes it encodes will then be expressed. This gene expression can be used to confirm successful conjugation.
For example, consider a scenario where the donor strain expresses ampicillin resistance and passes this on in the conjugated DNA to the recipient bacterium, but the recipient strain also has a tetracycline resistance gene not present in the donor. In this event, when the cells are plated on LB media containing both tetracycline and ampicillin, colonies should grow only from successfully conjugated bacteria, which will be expressing both resistance phenotypes. To further confirm successful conjugation, plasmid DNA from these colonies can be harvested and then a section of DNA specific to the transferred plasmid can be amplified using polymerase chain reaction, or PCR. When the PCR product is run on an electrophoresis gel alongside a ladder of standard sizes, a PCR fragment of a known size should be visible on the gel, further confirming successful conjugation. In this experiment, a plasmid will be used to transfer the ampicillin resistance gene via conjugation from a donor strain to a tetracycline-resistant recipient strain. After this, to confirm conjugation, the conjugation mixture will be incubated on a plate containing both antibiotics leaving only the transformed bacteria. Finally, successful conjugation will be further confirmed with PCR.
Before starting the procedure, put on the appropriate personal protective equipment, including a lab coat and gloves. Next, sterilize the workspace using 70% ethanol to wipe down the surface.
In this procedure, the ampicillin resistance gene will be transferred from the WM3064 strain of E. coli to the J53 strain of E. coli via conjugation. The donor strain WM3064 is resistant to tetracycline and ampicillin and it requires diaminopimelic acid, or DAP, to grow. The recipient strain J53 is only resistant to tetracycline and it does not require DAP to grow. This means that successfully conjugated cells should be resistant to tetracycline and ampicillin and can grow without DAP.
Prepare the donor strain culture by inoculating five milliliters of LB containing 0.3 millimoles of DAP with a scrap of the frozen donor strain glycerol stock. Then, prepare the recipient strain by inoculating five milliliters of LB broth without DAP with a scrap of the frozen recipient strain glycerol stock. Grow these cultures overnight at 37 degrees Celsius with aeration and shaking at 220 RPM in a shaking incubator. Once the cultures have grown to an OD 600 of two, remove one milliliter of culture from each and place this into two new separate 1.5 milliliter microcentrifuge tubes. Then, centrifuge these aliquots at 3000 RPM for five minutes to pellet the bacterial cells. Discard the supernatant and wash each pellet with 250 microliters of 1X PBS. Centrifuge the samples again and, after discarding the supernatant, resuspend each pellet in 500 microliters of PBS.
To begin the conjugation procedure, first combine 50 microliters of recipient cells with 50 microliters of donor cells in a 1.5 milliliter microcentrifuge tube and mix by pipetting up and down gently. Next, pipette 100 microliters of the recipient cell culture onto another 1X tetracycline plate containing DAP. Next, prepare your negative control by pipetting 100 microliters of the recipient cell culture only onto a non-selective agar plate containing DAP. Then, incubate the conjugation and negative control plates overnight at 37 degrees Celsius.
The next day, take a sterile cell scraper and harvest cells from the conjugation plate by collecting colonies. Then, transfer the colonies to a sterile 1.5 milliliter microcentrifuge tube containing one milliliter of 1X PBS. Repeat this process to collect the recipient cells from the other plate.
After this, vortex the samples to mix. After mixing, transfer the tubes to a centrifuge to gently pellet the cells. Discard the supernatant, then wash the cell pellets in one milliliter of PBS and vortex the tubes to resuspend the cells. Pellet the cells again by centrifuging. Discard the supernatant again and resuspend both cell pellets in one milliliter of PBS. Now, using a sterile pipette tip, plate 100 microliters of the conjugation reaction cell mixture onto an LB agar plate without DAP containing 1X tetracycline and 1X ampicillin. Repeat the plating method using 100 microliters of a ten-fold dilution of the same cell mixture in PBS onto another LB agar plate without DAP containing 1X tetracycline and 1X ampicillin.
Finally, pipette 100 microliters of the negative control cell mixture onto a single LB agar plate with 1X tetracycline only. After overnight incubation at 37 degrees Celsius, the colonies should be visible. Using a sterile pipette tip, pick a single colony from the conjugation reaction plate and add it to a tube containing five milliliters of selective LB media containing both antibiotics. Then, repeat the colony isolation by selecting a single colony from the recipient cell plate. Grow these cultures overnight at 37 degrees Celsius with aeration at 220 RPM.
The next day, wipe down the bench top with 70% ethanol and remove the plates from the incubator. Use a DNA mini prep kit to isolate DNA from 4. 5 milliliters of each culture according to the manufacturer’s instructions. After completing the DNA mini prep, elute the DNA using 35 microliters of nuclease-free water. Finally, use the remaining 0. 5 milliliters of each culture to prepare one milliliter glycerol stocks by adding 0.5 milliliters of 100% glycerol for a one-to-one dilution. Place these aliquots at minus 80 degrees Celsius for storage until needed.
To confirm successful conjugation by PCR, first prepare a PCR master mix by adding 75 microliters of 2X PCR master mix to a microcentrifuge tube. Then, add 7.5 microliters each of a 10 micromolar forward primer and a 10 micromolar reverse primer designed to amplify the ampicillin resistance gene from the plasmid. Next, prepare a second PCR master mix by adding 75 microliters of 2X PCR master mix to a microcentrifuge tube and then adding 7.5 microliters each of a 10 micromolar forward primer and 10 micromolar reverse primer designed to amplify a housekeeping gene, in this case DNA gyrase B.
Now, add 15 microliters of the first master mix to a PCR tube and then add 10 nanograms, approximately two microliters of the template experimental DNA to the same tube. Bring the reaction up to a final volume of 25 microliters with nuclease-free water. Repeat these steps to produce the remaining five reactions, so that the tubes contain the components shown here. Now, transfer these reactions to a thermocycler with the block pre-heated to 98 degrees Celsius and then initiate the program. After completion of the PCR, remove the tubes from the machine. Then, load two microliters of each reaction mixed with two microliters of loading dye and four microliters of a molecular weight marker into consecutive wells of a 1% agarose gel. Set the gel to run at 150 volts for 20 minutes. Finally, visualize the gel using a UV illuminator.
In this experiment, the successful transfer of the ampicillin resistance gene via conjugation was confirmed via PCR. Here, a roughly 500 base pair sized band should be observed in the well containing the conjugated DNA and ampicillin primers, well two in this example. A housekeeping gene, DNA gyrase B, was loaded into wells three and five with conjugated DNA and recipient cell DNA, respectively. Bands observed in these wells act as a positive control to ensure the DNA template was present and that PCR was successful. Bands should not be observed in the well containing the reaction for recipient cell DNA and the ampicillin primer pair, well four in this example, because the recipient cells are not ampicillin-resistant. Additionally, no bands should be observed in the reactions lacking template DNA, wells six and seven here. If these conditions are met, this will confirm the successful transfer of the ampicillin resistance gene, conferring ampicillin resistance from the WM3064 strain of E. coli to the J53 strain of E. coli.
