접합: 공여 E.coli에서 수용 E.coli로 암피실린 내성을 전달하는 방법
English
Share
Overview
출처: 알렉산더 S. 골드1, 토냐 M. 콜핏스1
1 미생물학학과, 보스턴 대학교 의과대학, 국립 신흥 감염 질병 연구소, 보스턴, MA
1946년 Lederberg와 테이텀에 의해 처음 발견된, 결합은 두 개의 세균세포(1)의 직접적인 물리적 접촉에 의존하는 박테리아 간의 수평 유전자 전달의 한 형태이다. 변형 이나 트랜스포션과 같은 다른 형태의 유전자 전달과 달리, 컨쥬게이션은 DNA가 기증자 세포에서 단방향 방식으로 받는 세포로 분비되는 자연발생 적인 과정입니다. 이러한 방향성 및 박테리아의 유전적 다양성을 증가시키는 이 과정에 대한 능력은 최근 항생제 내성 박테리아(2, 3)의 상승에 크게 기여한 것으로 여겨지는 세균성 “짝짓기”의 한 형태로 명성을 얻었습니다. 선택적 압력을 이용하여, 예를 들어 항생제의 사용, 결합은 실험실 환경에서 사용하기 위해 조작되어 박테리아 간의 수평 유전자 전달을 위한 강력한 도구이며, 어떤 경우에는 박테리아에서 효모, 식물 및 동물 세포(4)로 균계로 이동한다. 실험실에서 응용 프로그램 외에도, 균-진핵생물 유전자 전달 은 가능한 생체 공학 응용 및 자연 발생 의미 (5)의 무리와 DNA 전송의 흥미로운 경로입니다.
컨쥬게이션은 “2단계 메커니즘”(6)에 의해 작동하도록 생각됩니다. 첫째, 어떤 DNA든지 전송될 수 있기 전에, 기증자 세포는 수령인과 직접 세포 대 세포 접촉을 해야 합니다. 이 과정은 그람 음성 박테리아에서 가장 잘 특징지어졌으며, 가장 많이 연구된 것은 대장균이다. 세포-세포 접촉은 F(불임) 인자(7, 8)로 알려진 전달 가능한 유전자에 의해 인코딩된 유도체인 성필루스로 알려진 기증자에 세포외 필라멘트의 복잡한 네트워크의 존재에 의해 확립된다. 기증자와 수령인 간의 접촉을 확립하는 것 외에도, 여러 단백질은 성필루스를 통해 받는 자 세포질로 이송되어 두 세포 사이에 형 IV 분비 시스템(T4SS) 도관을 형성하며, 2차 컨쥬게이션, DNA 전달(6)에 필요한 구조이다. 성필루의 이러한 기능을 DNA의 롤링 원 복제와 결합함으로써, 기증자 세포는 플라스미드 또는 트랜스포슨과 같은 형질전환 가능한 요소의 형태로 DNA를 “촬영 및 펌프” 모델(6)에 의해 받는 사람에게 전달할 수 있다. 이 경우, “사격”은 시험 단백질의 수송이며, 연결된 DNA를 가진, T4SS에 의해 수신자 세포로, 그리고 “펌핑”은 수신자에게 DNA의 활성 수송, T4SSS에 의존하는 과정및 결합 단백질(6)에 의해 촉매된다. 이 과정에 사용되는 기계는 시스 및 트랜스 유전자의 DNA에 의해 제공되어야 하는 전사 서열(oriT)의 기원으로 구성되며, 이는 이완제, 짝짝 쌍 형성 복합체 및 타입 IV 커플링 단백질을 인코딩하고 시스 또는 트랜스(9)에 존재할 수 있다. 이 릴렉스아제는 오리T 서열 내에서 닉 사이트를 갈라놓고 다른 보조 단백질 (9)과 함께 단일 가닥 DNA 이완제 복합체인 이완을 생성하기 위해 전달된 가닥의 5’말에 공유적으로 부착합니다. 일단 형성되면, 이완성은 T4SS (10)에 의해 수신자 세포로 ssDNA-relaxase 복합체의 전송을 허용하는 타입 IV 커플링 단백질을 통해 결합 쌍 형성 복합체에 연결합니다. 일단 수령인의 세포질에서, DNA는 수신자 게놈으로 통합하거나 그것의 유전자의 발현을 허용하는 플라스미드의 양식에서 별도로 존재할 수 있습니다.
본 실험에서, 널리 사용되는 유도균 균주 대장균 WM3064는 수취인균 균주 E. coli J53에 ampicillin 내성을 위한 유전자 인코딩을 전송하는 데 사용되었다. 그람 음성 박테리아의 두 변종은 테트라사이클린에 저항하는 동안, 만 기증자 균주 WM3064는 ampicillin 저항을 위한 유전자를 가지고, pWD2-oriT 셔틀 벡터에 대한 인코딩, 및 diaminopimelic 산 (DAP)에 보조 trophic이었다 (11-13). 본 실험은 두 가지 주요 단계, 기증자 및 수령인 균주의 제제, 유도체에서 양수로 암피실린 저항 유전자의 전송으로 구성하였다(도 1).
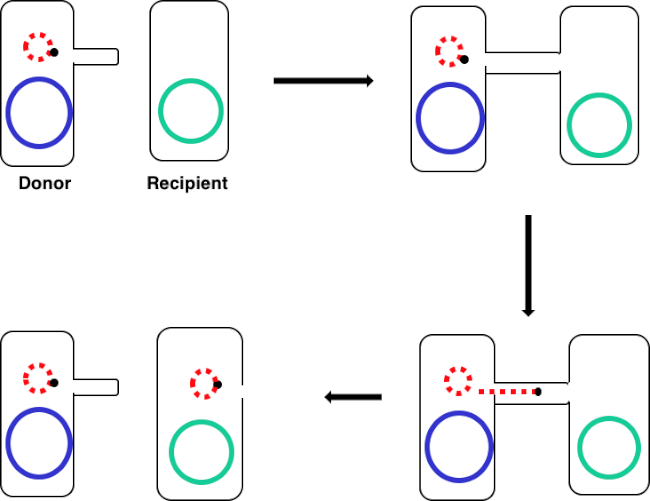
그림 1: 컨쥬게이션 회로도. 이 회로도는 골치를 이용한 기증자 세포에서 수신자 세포로 의한 전형 가능한 DNA 요소의 단 하나의 예인 플라스미드의 성공적인 전달을 보여줍니다. 성별 필러스를 통해 기증자 세포에 의해 받는 사람 세포와 접촉하면, 플라스미드는 롤링 원 복제에 의해 복제되고, 두 세포에 결합하는 다단백 복합체를 통해 이동하고, 받는 사람 세포에 새로운 전체 길이 플라스미드를 형성한다.
기증자와 수령인 세포의 혼합물을 배양함으로써 테트라사이클린과 DAP의 존재속에서 이러한 세포를 연속적으로 도금함으로써, 이것은 암피실린 저항 유전자의 성공적인 전달을 허용했다. 테트라사이클린과 암피실린이 존재하여 이 혼합물에서 자라는 세포는, 증폭균 저항 유전자를 얻지 못했을 수 있는 DAP 및 수신자 세포의 부족으로 인해 모든 기증자 세포를 제거하여 암피실린 저항성을 획득한 J53 균주 균을 엄격하게 산출하였다(도 2). 일단 수행되면, 암피실린 저항 유전자의 성공적인 전송은 PCR에 의해 확인되었다. 인쥬게이션이 성공했기 때문에 대장균의 J53 균주는 pWD2-oriT를 함유하고 암피실린에 내성이 있었으며, 이러한 저항을 위한 유전자 인코딩은 PCR에 의해 검출될 수 있다. 그러나, 성공하지 못하면 암피실린 저항 유전자의 검출이 없었을 것이고 암피실린은 여전히 J53 균주에 대한 효과적인 항생제로 기능할 것이다.
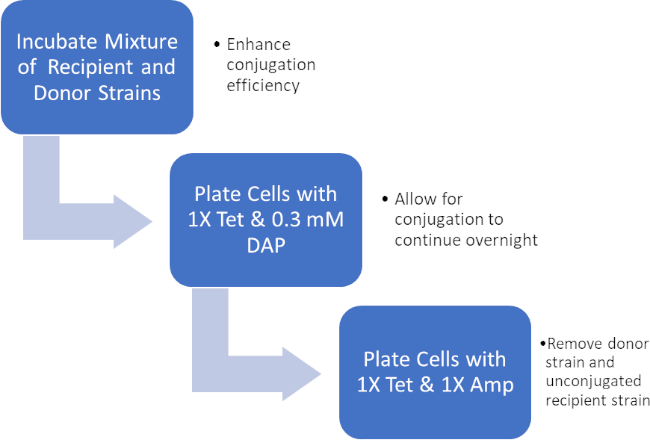
그림 2: 프로토콜 회로도. 이 회로도는 제시된 프로토콜에 대한 개요를 보여 주어 있습니다.
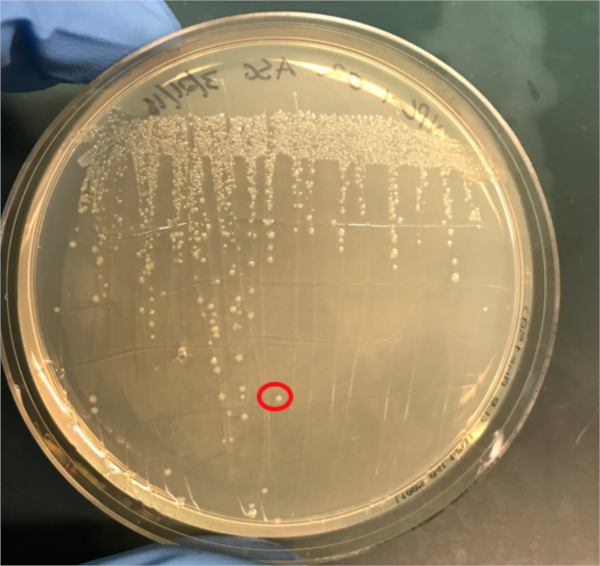
그림 3A: PCR에 의한 성공적인 컨쥬게이션의 확인. A) 컨쥬게이트 및 음극식 샘플의 냉동고 주식이 천판에 줄지어 서 있었고, DNA 절연을 위해 콜로니가 선택되었다(빨강).
Procedure
Results
If conjugation was successful, a 500 base-pair sized band PCR product will be observed in the well in which PCR reaction 1 was loaded (Well #2 in Figure 3B), while no bands will be observed in the well in which PCR reaction 3 was loaded (Well #4 in Figure 3B). The presence of this band confirms the successful transfer of the ampicillin resistance gene, thereby conferring ampicillin resistance to the J53 strain of E. coli.
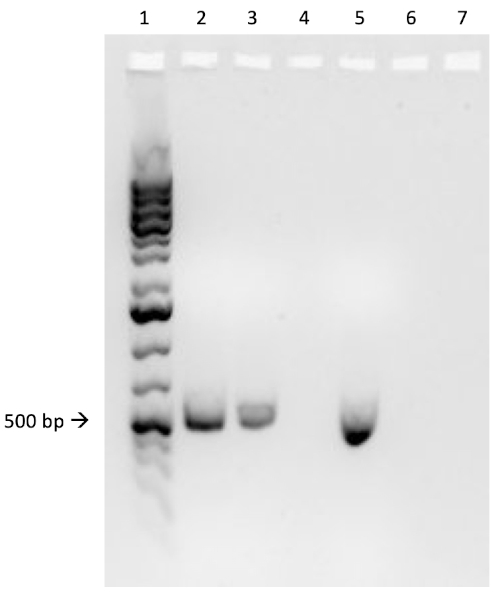
Figure 3B: The confirmation of successful conjugation by PCR. B) PCR analysis was done using DNA isolated from the select colony. The contents of each well are as follows: 1) DNA ladder, 2) Conjugation DNA and ampicillin primers, 3) Conjugation DNA and housekeeping primers, 4) Negative control DNA and ampicillin primers, 5) Negative control DNA and housekeeping primers, 6) No DNA and ampicillin primers, and 7) No DNA and negative control primers. The presence of a ~ 500 base-pair band PCR product from PCR reaction 1 (well 2), and the lack of this product from PCR reaction 3 (well 4), confirms successful conjugation.
Applications and Summary
Conjugation is a naturally occurring process of horizontal gene transfer that relies on the direct cell-to-cell contact of a donor cell and a recipient cell. This process is shared among all kinds of bacteria and has been instrumental in bacterial evolution, most notably antibiotic resistance. In the lab, conjugation can be used as an effective method of gene transfer that is much less disruptive when compared to other techniques. Outside of the laboratory, the ability to transfer DNA from bacteria to eukaryotes via conjugation offers an exciting new avenue of gene therapy and understanding the implications of these naturally occurring gene transfers, for example the relationship between bacterial infection and cancer, is a rapidly emerging area of research.
References
- Lederberg J, Tatum, E.L. Gene recombination in Escherichia coli Nature. 1946;158:558.
- Holmes R.K. J, M.G. Genetics: Exchange of Genetic Information. 4th Edition ed. Baron S, editor. Galveston, TX: University of Texas Medical Branch at Galveston; 1996.
- Cruz F, Davies, J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends in Microbiology. 2000;8:128-33.
- Llosa M, Cruz, F. Bacterial conjugation: a potential tool for genomic engineering. Ressearch in Microbiology. 2005;156:1-6.
- Lacroix B, Citovsky, V. Transfer of DNA from Bacteria to Eukaryotes. mBio. 2016;7(4):1-9.
- Llosa M, et al. Bacterial conjugation: a two-step mechanism for
- DNA transport. Molecular Microbiology. 2002;45:1-8.
- Grohmann E, Muth, G., Espinosa, M. Conjugative Plasmid Transfer in Gram-Positive Bacteria. Microbiology and Molecular Biology Reviews. 2003;67:277-301.
- Firth N, Ippen-Ihler, K, Skurray, RA. Structure and function of the F factor and mechanism of conjugation. Escherichia coli and salmonella: cellular and molecular biology. 1996;2:2377-401.
- Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EPC, De La Cruz F. Mobility of Plasmids. Microbiology and Molecular Biology Reviews. 2010;74(3):434-52.
- Cascales E. Definition of a Bacterial Type IV Secretion Pathway for a DNA Substrate. 2004;304(5674):1170-3.
- Wang P, Yu Z, Li B, Cai X, Zeng Z, Chen X, et al. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Microbial Cell Factories. 2015;14(1):11.
- Yi H, Cho YJ, Yong D, Chun J. Genome Sequence of Escherichia coli J53, a Reference Strain for Genetic Studies. Journal of Bacteriology. 2012;194(14):3742-3.
- Baumann RLB, E. H.; Wiseman, J. S.; Vaal, M.; Nichols, J. S. Inhibition of Escherichia coli Growth and Diaminopimelic Acid Epimerase by 3-Chlorodiaminopimelic Acid. Antimicrobial Agents and Chemotherapy 1988;32:1119-23.
- Rocha D, Santos, CS, Pacheco LG. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek. 2015;108:685-93.
Transcript
Bacterial cells, such as E. coli, are able to transfer genetic information from cell-to-cell. Conjugation differs from other mechanisms of DNA transfer, such as transduction or transformation, in that it requires physical contact between the cells.
To proceed, conjugation requires a donor cell that expresses the fertility, or F, factor and a recipient cell without it, an F minus cell. The process requires two steps. The first is the establishment of direct cell-to-cell contact. To do this, the donor cell generates an extracellular filamentous structure called a sex pilus. It is named this since conjugation is a form of mating for asexually reproducing bacteria, but it should be noted that it is not true sexual reproduction as no gametes are exchanged and no offspring are formed.
The second step is delivery of DNA to the recipient cell. After the sex pilus establishes contact between two cells, a conduit called the Type IV secretion system is built allowing for the transfer of DNA. The donor cell then begins to replicate the extrachromosomal DNA that will be transferred selected based on the presence of a genetic element known as the OriT or origin of transfer. One end of the newly replicated DNA is threaded into the conduit through DNA protein binding. As the DNA is further replicated, it is pumped through the channel, facilitated by a complex of proteins encoded by genes located close to the OriT. Once the DNA is fully transferred, it will either form an extra chromosomal plasmid, or it may integrate into the chromosome of the recipient cell. Whichever the endpoint of the transferred DNA, the genes it encodes will then be expressed. This gene expression can be used to confirm successful conjugation.
For example, consider a scenario where the donor strain expresses ampicillin resistance and passes this on in the conjugated DNA to the recipient bacterium, but the recipient strain also has a tetracycline resistance gene not present in the donor. In this event, when the cells are plated on LB media containing both tetracycline and ampicillin, colonies should grow only from successfully conjugated bacteria, which will be expressing both resistance phenotypes. To further confirm successful conjugation, plasmid DNA from these colonies can be harvested and then a section of DNA specific to the transferred plasmid can be amplified using polymerase chain reaction, or PCR. When the PCR product is run on an electrophoresis gel alongside a ladder of standard sizes, a PCR fragment of a known size should be visible on the gel, further confirming successful conjugation. In this experiment, a plasmid will be used to transfer the ampicillin resistance gene via conjugation from a donor strain to a tetracycline-resistant recipient strain. After this, to confirm conjugation, the conjugation mixture will be incubated on a plate containing both antibiotics leaving only the transformed bacteria. Finally, successful conjugation will be further confirmed with PCR.
Before starting the procedure, put on the appropriate personal protective equipment, including a lab coat and gloves. Next, sterilize the workspace using 70% ethanol to wipe down the surface.
In this procedure, the ampicillin resistance gene will be transferred from the WM3064 strain of E. coli to the J53 strain of E. coli via conjugation. The donor strain WM3064 is resistant to tetracycline and ampicillin and it requires diaminopimelic acid, or DAP, to grow. The recipient strain J53 is only resistant to tetracycline and it does not require DAP to grow. This means that successfully conjugated cells should be resistant to tetracycline and ampicillin and can grow without DAP.
Prepare the donor strain culture by inoculating five milliliters of LB containing 0.3 millimoles of DAP with a scrap of the frozen donor strain glycerol stock. Then, prepare the recipient strain by inoculating five milliliters of LB broth without DAP with a scrap of the frozen recipient strain glycerol stock. Grow these cultures overnight at 37 degrees Celsius with aeration and shaking at 220 RPM in a shaking incubator. Once the cultures have grown to an OD 600 of two, remove one milliliter of culture from each and place this into two new separate 1.5 milliliter microcentrifuge tubes. Then, centrifuge these aliquots at 3000 RPM for five minutes to pellet the bacterial cells. Discard the supernatant and wash each pellet with 250 microliters of 1X PBS. Centrifuge the samples again and, after discarding the supernatant, resuspend each pellet in 500 microliters of PBS.
To begin the conjugation procedure, first combine 50 microliters of recipient cells with 50 microliters of donor cells in a 1.5 milliliter microcentrifuge tube and mix by pipetting up and down gently. Next, pipette 100 microliters of the recipient cell culture onto another 1X tetracycline plate containing DAP. Next, prepare your negative control by pipetting 100 microliters of the recipient cell culture only onto a non-selective agar plate containing DAP. Then, incubate the conjugation and negative control plates overnight at 37 degrees Celsius.
The next day, take a sterile cell scraper and harvest cells from the conjugation plate by collecting colonies. Then, transfer the colonies to a sterile 1.5 milliliter microcentrifuge tube containing one milliliter of 1X PBS. Repeat this process to collect the recipient cells from the other plate.
After this, vortex the samples to mix. After mixing, transfer the tubes to a centrifuge to gently pellet the cells. Discard the supernatant, then wash the cell pellets in one milliliter of PBS and vortex the tubes to resuspend the cells. Pellet the cells again by centrifuging. Discard the supernatant again and resuspend both cell pellets in one milliliter of PBS. Now, using a sterile pipette tip, plate 100 microliters of the conjugation reaction cell mixture onto an LB agar plate without DAP containing 1X tetracycline and 1X ampicillin. Repeat the plating method using 100 microliters of a ten-fold dilution of the same cell mixture in PBS onto another LB agar plate without DAP containing 1X tetracycline and 1X ampicillin.
Finally, pipette 100 microliters of the negative control cell mixture onto a single LB agar plate with 1X tetracycline only. After overnight incubation at 37 degrees Celsius, the colonies should be visible. Using a sterile pipette tip, pick a single colony from the conjugation reaction plate and add it to a tube containing five milliliters of selective LB media containing both antibiotics. Then, repeat the colony isolation by selecting a single colony from the recipient cell plate. Grow these cultures overnight at 37 degrees Celsius with aeration at 220 RPM.
The next day, wipe down the bench top with 70% ethanol and remove the plates from the incubator. Use a DNA mini prep kit to isolate DNA from 4. 5 milliliters of each culture according to the manufacturer’s instructions. After completing the DNA mini prep, elute the DNA using 35 microliters of nuclease-free water. Finally, use the remaining 0. 5 milliliters of each culture to prepare one milliliter glycerol stocks by adding 0.5 milliliters of 100% glycerol for a one-to-one dilution. Place these aliquots at minus 80 degrees Celsius for storage until needed.
To confirm successful conjugation by PCR, first prepare a PCR master mix by adding 75 microliters of 2X PCR master mix to a microcentrifuge tube. Then, add 7.5 microliters each of a 10 micromolar forward primer and a 10 micromolar reverse primer designed to amplify the ampicillin resistance gene from the plasmid. Next, prepare a second PCR master mix by adding 75 microliters of 2X PCR master mix to a microcentrifuge tube and then adding 7.5 microliters each of a 10 micromolar forward primer and 10 micromolar reverse primer designed to amplify a housekeeping gene, in this case DNA gyrase B.
Now, add 15 microliters of the first master mix to a PCR tube and then add 10 nanograms, approximately two microliters of the template experimental DNA to the same tube. Bring the reaction up to a final volume of 25 microliters with nuclease-free water. Repeat these steps to produce the remaining five reactions, so that the tubes contain the components shown here. Now, transfer these reactions to a thermocycler with the block pre-heated to 98 degrees Celsius and then initiate the program. After completion of the PCR, remove the tubes from the machine. Then, load two microliters of each reaction mixed with two microliters of loading dye and four microliters of a molecular weight marker into consecutive wells of a 1% agarose gel. Set the gel to run at 150 volts for 20 minutes. Finally, visualize the gel using a UV illuminator.
In this experiment, the successful transfer of the ampicillin resistance gene via conjugation was confirmed via PCR. Here, a roughly 500 base pair sized band should be observed in the well containing the conjugated DNA and ampicillin primers, well two in this example. A housekeeping gene, DNA gyrase B, was loaded into wells three and five with conjugated DNA and recipient cell DNA, respectively. Bands observed in these wells act as a positive control to ensure the DNA template was present and that PCR was successful. Bands should not be observed in the well containing the reaction for recipient cell DNA and the ampicillin primer pair, well four in this example, because the recipient cells are not ampicillin-resistant. Additionally, no bands should be observed in the reactions lacking template DNA, wells six and seven here. If these conditions are met, this will confirm the successful transfer of the ampicillin resistance gene, conferring ampicillin resistance from the WM3064 strain of E. coli to the J53 strain of E. coli.
