Real-time Iontophoresis with Tetramethylammonium to Quantify Volume Fraction and Tortuosity of Brain Extracellular Space
Summary
This protocol describes real-time iontophoresis, a method that measures physical parameters of the extracellular space (ECS) of living brains. The diffusion of an inert molecule released into the ECS is used to calculate the ECS volume fraction and tortuosity. It is ideal for studying acute reversible changes to brain ECS.
Abstract
This review describes the basic concepts and protocol to perform the real-time iontophoresis (RTI) method, the gold-standard to explore and quantify the extracellular space (ECS) of the living brain. The ECS surrounds all brain cells and contains both interstitial fluid and extracellular matrix. The transport of many substances required for brain activity, including neurotransmitters, hormones, and nutrients, occurs by diffusion through the ECS. Changes in the volume and geometry of this space occur during normal brain processes, like sleep, and pathological conditions, like ischemia. However, the structure and regulation of brain ECS, particularly in diseased states, remains largely unexplored. The RTI method measures two physical parameters of living brain: volume fraction and tortuosity. Volume fraction is the proportion of tissue volume occupied by ECS. Tortuosity is a measure of the relative hindrance a substance encounters when diffusing through a brain region as compared to a medium with no obstructions. In RTI, an inert molecule is pulsed from a source microelectrode into the brain ECS. As molecules diffuse away from this source, the changing concentration of the ion is measured over time using an ion-selective microelectrode positioned roughly 100 µm away. From the resulting diffusion curve, both volume fraction and tortuosity can be calculated. This technique has been used in brain slices from multiple species (including humans) and in vivo to study acute and chronic changes to ECS. Unlike other methods, RTI can be used to examine both reversible and irreversible changes to the brain ECS in real time.
Introduction
The extracellular space (ECS) is the network of interconnected channels exterior to all brain cells and contains both interstitial fluid and extracellular matrix (Figure 1a and Figure 1b). The distribution of many substances required for brain cell function, including nutrients, hormones, and neurotransmitters, occurs by diffusion through the ECS. Changes in the physical parameters of this space, including volume, geometry, and extracellular matrix, can drastically affect diffusion through the ECS and the local ion concentrations bathing brain cells, which have a profound impact on brain cell function1,2.
Real-time iontophoresis (RTI) is used to determine two structural characteristics of a brain region: volume fraction and tortuosity3,4,5. Volume fraction (α) is the proportion of tissue volume occupied by the ECS (VECS) relative to the total tissue volume (Vtissue) in a representative elementary volume;
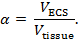
Tortuosity (λ) is the relative hindrance that a substance encounters when diffusing through a brain region as compared to a medium with no obstructions;
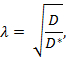
where D* (cm2 s-1) is the effective diffusion coefficient of the substance in brain and D (cm2s-1) is the free diffusion coefficient of the substance in a free medium, such as dilute agarose gel.
Today, the most commonly used probe substance for the RTI method is the small cation tetramethylammonium (TMA). TMA has a molecular weight of 74 g/mol, completely dissociates in solution, and has one positive charge. RTI studies with this ion have demonstrated that α 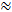 0.2 and λ
0.2 and λ 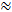 1.61,2. This means that the ECS is roughly 20% of the total brain volume and that the diffusion of a small, inert molecule occurs roughly 2.5 times slower in the ECS than in a medium with no obstructions3. However, both α and λ vary with brain age, region, and state and in pathological conditions1. Alterations of these parameters have been linked to brain development, aging, sleep, epilepsy, and many other fundamental processes and diseases of the brain1,6. While other techniques measure α and λ, RTI can measure both in localized regions of living tissue in real time. For this reason, RTI has become an indispensable tool for investigating changes in α and λ during acute and reversible challenges.
1.61,2. This means that the ECS is roughly 20% of the total brain volume and that the diffusion of a small, inert molecule occurs roughly 2.5 times slower in the ECS than in a medium with no obstructions3. However, both α and λ vary with brain age, region, and state and in pathological conditions1. Alterations of these parameters have been linked to brain development, aging, sleep, epilepsy, and many other fundamental processes and diseases of the brain1,6. While other techniques measure α and λ, RTI can measure both in localized regions of living tissue in real time. For this reason, RTI has become an indispensable tool for investigating changes in α and λ during acute and reversible challenges.
The theory supporting RTI was originally validated by Nicholson and Phillips, and the technique has been used extensively since that time4,7. Experiments employing RTI begin with the release of a pulse of TMA from a source microelectrode by iontophoresis into a dilute agarose gel. Once ejected, the ions freely diffuse away from the point source, choosing from a potentially infinite number of random paths (Figure 1d). The changing concentration of the ion is measured over time using an ion-selective microelectrode (ISM) positioned roughly 100 µm away (Figure 1c). The changes in TMA concentration are graphed and fitted to a curve that allows for the calculation of both D and the transport number of the iontophoresis microelectrode (parameters discussed in the Protocol). With these values, the procedure is repeated in a brain region of interest to obtain D* and to calculate both α and λ. Control of the iontophoresis microelectrode, data collection, graphing and fitting of the TMA concentration curve, and calculation of the experimental parameters are all typically done by the programs Wanda and Walter, which have been specifically designed for this purpose (the software and their manuals are freely available from the authors upon request).
The Protocol section of this review describes the basic procedures needed to design and perform RTI in rodent brain slices. The technique has also been used in non-rodent models, including human brain slices and in vivo brain preparations1,4,6,8,9. The Representative Results section provides both ideal and non-ideal results to highlight nuances in data interpretation. Finally, the Discussion section briefly covers troubleshooting techniques, limitations of RTI, alternative techniques used to study the ECS, and future applications of RTI.
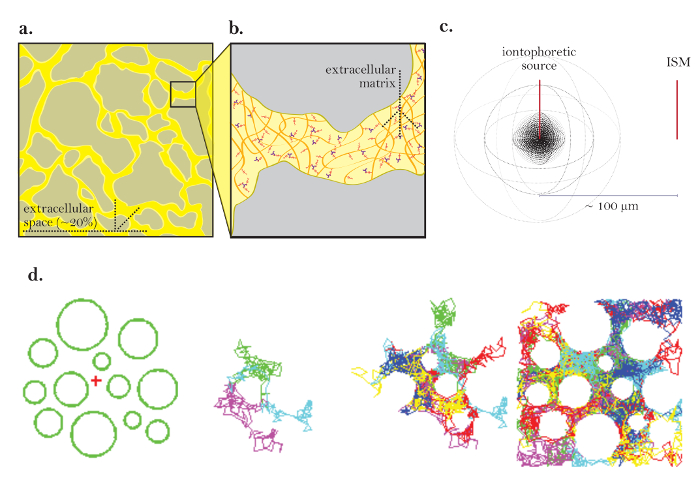
Figure 1: Diagrams of Diffusion through ECS. (a) Diagram of ECS: Demonstrates the size and location of the ECS in a typical brain section. Yellow marks the ECS between the gray brain cell processes. The volume of the ECS is roughly 20% of the total tissue volume (i.e., volume fraction = 0.2) under physiological conditions. (b) Magnified diagram of the ECS: Highlights physical parameters contributing to tortuosity, including brain cell geometry (gray) and extracellular matrix (diagramed as a mesh of multicolored glycosaminoglycans and proteoglycans). (c) 3D diagram of diffusion from a point source: Demonstrates the net movement of inert molecules from an iontophoretic source to an ISM. Excluding diffusion barriers and cellular uptake, molecules diffuse outwards in all directions, producing a spherical concentration front. The ISM quantifies the local concentration of the inert molecules released from the iontophoretic source. (d) Computer simulation of diffusion in ECS of brain: [Far left] Setup for Monte Carlo simulation; green spheres represent brain cell processes and the red cross represents a point source. This setup models the brain tissue diagrammed in Figure 1a. [Middle images] 3 and 6 molecules performing random movements as they diffuse through the extracellular space of the brain, shown in 2 dimensions. [Far right] Random walks of many molecules released from the point source. The net movement of all molecules from the point source is outwards as depicted in Figure 1c. The cumulative random walks outline the spaces between the cells (i.e., the ECS; see reference5 for further explanation). Please click here to view a larger version of this figure.
Protocol
All animal procedures, used to obtain tissue samples, were approval by the animal ethics committee at SUNY Downstate Medical Center.
1. Preparation of Solutions and Equipment
- Prepare a 150 mM NaCl backfill solution for the reference barrel of the ISM. Store it in a 10 mL syringe attached to a 0.22 µm filter (to remove bacteria or particles).
- Prepare a 150 mM TMA chloride (TMA-Cl) backfill solution for the microelectrodes. Store it in a 10 mL syringe attached to a 0.22 µm filter. Prepare the TMA-Cl solutions (in this protocol) from a 5 M manufacturer stock solution to ensure the correct concentration.
- Chloridize at least four silver wires for the fabrication of microelectrodes by submerging the wires in bleach (sodium hypochlorite) for at least 2 h. Remove excess bleach with ethanol and allow the wires to dry.
- Prepare 50 mL of 0.3% agarose in 150 mM NaCl and 0.5 mM TMA-Cl in a beaker and cover it. Use agarose that is powdered and reasonably fresh to ensure good diffusion measurements.
- Heat and mix the agarose solution with a stir bar to dissolve it. Allow the solution to cool to room temperature. Store this at 4 °C for up to 1 week.
- Prepare an indifferent (ground) electrode made out of 4% agarose in 1 M KCl (directions in Supplement A)
- Fabricate a small, porous cup that can fit in the experimental chamber and that allows for electrical continuity between its contents and the outside environment (Figure 2a). Place a metal ring on the bottom of this cup to prevent it from floating when partially submerged in water.
- Use serial dilution of a 5 M TMA-Cl stock to make five 100 mL TMA-Cl solutions for the calibration of the ISMs. Solutions should have final concentrations of 0.5, 1, 2, 4, and 8 mM TMA-Cl, all in 150 mM NaCl. Store the calibration solutions in a sealable cup to prevent evaporation.
2. Electronic Setup
- Connect the components of the RTI experimental setup according to the block diagram in Figure 2b; include an amplifier with two input channels (one of which should be very high-impedance for the ion-selective barrel of the ISM), a low-pass filter set at 10 Hz, a chart recorder, an A/D + D/A converter, an iontophoretic unit (or an amplifier capable of supplying constant-current pulses), and a computer (PC) running the Wanda and Walter programs. Inspect the electronic setup to confirm that all connections are in place.
- Shield the experimental setup in a grounded enclosure (such as a Faraday cage), if necessary, as ISMs have a high resistance and are sensitive to artifacts created by nearby movement.
- Create a dedicated ISM calibration station consisting of a dual input amplifier, a chart recorder, an appropriate ISM holder, and an indifferent ground electrode. If possible, shield the enclosure. Skip this step if the ISMs are calibrated in the experimental setup (step 3.29)
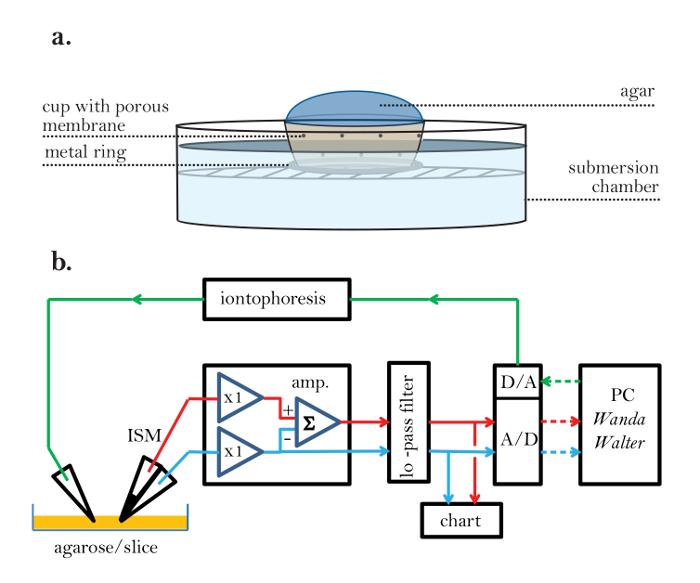
Figure 2: Porous Experimental Cup and Electronic Setup. (a) Porous experimental cup: A porous mesh is used to create an experimental cup that allows for electrical continuity between the agarose (inside) and the experimental bathing fluid (outside). A metal ring is attached to the bottom of the cup to prevent the cup from floating in the bathing solution. (b) Block diagram of the RTI setup (steps 2.1 and 2.2): An ISM is connected to an amplifier (amp.). The ISM has two barrels. One contains liquid ion exchanger (LIX) in the tip and generates a voltage proportional to the logarithm of the TMA concentration at the tip together with the local ambient voltage; the signal path is represented by a red line. The other barrel of the ISM is known as the reference barrel and measures the ambient voltage at the tip of the ISM; it is connected by a blue signal path. The amplifier has two so-called head stages that connect to the ISM; these units have a gain of 1 (x1) and match the high impedance of the microelectrode to the low impedance of the rest of the amplifier circuitry. The head stage connected to the ion-selective barrel must be able to match an incoming resistance of about 1,000 MΩ, whereas the resistance of the reference barrel is typically about 10 MΩ. After leaving the head stage, the voltage from the reference barrel is inverted and subtracted from the voltage on the ion-selective barrel using a summing amplifier (Σ) to obtain the pure ion signal voltage. The outputs of the amplifier pass to a signal conditioning unit that provides additional amplification and a multipole low-pass filter (≤10 Hz; typically a Bessel filter), which removes noise and prevents signal aliasing at the analog-to-digital converter (A/D). The outputs of the filter are also displayed on a strip chart recorder. The A/D converter digitizes the signals and sends them to a personal computer (PC). The PC also generates a digital signal that is converted by a digital-to-analog converter (D/A) to an analog voltage pulse that is fed to the iontophoresis unit, which converts the voltage to a current pulse of constant amplitude and sends it to the iontophoresis microelectrode. The iontophoresis signal path is represented by a green line. The data acquisition and iontophoresis signal are under the control of the Wanda program, which generates an output file for each diffusion record in the form of a voltage versus time recording, along with all the parameters that define the experiment. A second program, Walter, reads the output file and uses ISM calibration data to convert the digitized voltages to concentrations. The concentration versus time curves are then fitted in Walter to the appropriate solution to the diffusion equation. D and nt are extracted if the medium is agarose, and λ and α extracted if the medium is brain. Analog signals are solid lines; digital signals are dotted lines. There is also an indifferent ground electrode (not shown) in the bath containing the slice. Red lines = ion signal, Blue lines = reference signal, Green lines = iontophoresis command, Solid lines = analog, Dotted lines = digital. Please click here to view a larger version of this figure.
3. Preparation and Calibration of Ion-selective Microelectrodes
- Fabricate ISMs using the protocol below one day prior to the experiment. Make ISMs in batches to ensure that at least two work on the day of the experiment.
NOTE: Most ISMs are stable for a day or two. ISM fabrication is sensitive to humidity and atmospheric conditions. Not every microelectrode will calibrate successfully. - Chip away roughly 0.5 cm of glass at the end of one of the barrels of a double-barreled borosilicate glass capillary using an old pair of forceps.
- Chip a single barrel on the opposite end of the capillary (Figure 3a). Ensure that the septum is not damaged (critical). Caution: Wear goggles to prevent injury due to projectile glass.
- Place the capillary in a bottle of acetone for at least 1 h to remove contaminants.
- Remove the capillary from the acetone and pulse clean, dry, compressed nitrogen gas or air through it to remove any excess acetone. Remove all acetone in the capillary, as residual acetone can interfere with silanization (crucial).
- Fabricate the tip of the micropipette on either a vertical or horizontal puller. Tailor the parameters to pull a pipette with a long taper and sharp tip, about 1 µm or less in diameter. At the end of this step, one capillary will be made into two pipettes (Figure 3a).
- Visualize a single micropipette under a compound, upright microscope with a 10X objective. Cut the tip off using a glass microscope slide so that the final diameter of the tip (i.e., both barrels) is between 2 and 5 µm (Figure 3b). This pipette will be referred to as an ISM from now on.
- Fill the chipped barrel of the ISM with 150 mM NaCl reference solution through the opening on the chipped side using a 10 mL syringe attached to a 0.22 µm filter and a 28 G, 97 mm needle (Figure 3b). Do not fill the barrel past three-fourths the height of the barrel.
- Fill the non-chipped barrel of the ISM with 150 mM TMA-Cl backfill solution. Tap the ISM gently to knock any air bubbles out of the solution. Check for bubbles under the microscope used for chipping the tip.
- Flame the back of the ISM using a Bunsen burner to ensure that no communication of the backfill solution occurs across the septum at the back of the ISM. Ensure that the top one-fourth of the ISM is dry after flaming.
- Insert a chloridized silver wire into the reference solution of the ISM and bend the wire protruding from the capillary to mark this as the reference barrel (Figure 3c). Ensure that the wire is submerged in the backfill solution and remains in solution for the duration of the experiment.
- Slide a short length of polytetrafluoroethylene tubing (about 20 cm long) over the tip of a 25 G syringe needle. Place the other end of the tubing in the back of the ion-selective barrel. Ensure that the tubing is in the barrel but above the backfill solution (Figure 3c).
- Heat a stick of dental wax with a Bunsen burner and seal both the tubing and the silver wire into their respective barrels (Figure 3c). Ensure that a complete air seal is produced around the plastic tubing in the ion-selective barrel (critical).
- Prepare a small, transparent glass container (5 mL or less) of 4% chlorotrimethylsilane in xylene. Caution: Xylenes and silanes are very hazardous to health; handle both chemicals inside a fume hood and discard appropriately.
- Position the container in front of a stereo dissection microscope mounted horizontally in a fume hood. Secure the ISM vertically over the container using a micromanipulator (Figure 3d).
- Dip the tip of the microelectrode in the chlorotrimethylsilane solution.
- Attach an empty 10-mL syringe to the 25-gauge needle leading to the ISM. Apply positive air pressure from the syringe until a bubble of TMA-Cl solution is formed; this step should be performed under direct visualization through the microscope.
- Tap the ISM holder gently to knock the bubble off of the tip.
- Draw the chlorotrimethylsilane solution to a height of roughly 1,500 µm into the tip of the ISM using negative pressure on the 10 mL syringe.
- Completely expel the chlorotrimethylsilane solution from the tip of the ISM until a bubble of TMA-Cl solution is created at the tip (Figure 3d).
- Repeat steps 3.19 and 3.20 five times. Ensure that an even, uninterrupted column of fluid is drawn into the tip each time. If no solution can be drawn into the tip, check whether the tubing is blocked, the air seal is incomplete, or the tip of the ISM is blocked.
- Flush all of the chlorotrimethylsilane solution out of the tip until a bubble of TMA-Cl solution is created.
- While maintaining positive pressure on the syringe, remove the ISM from the xylene solution. Ensure that all the xylene solution is expelled from the ISM tip, as excess xylene will ruin the exchanger column created in subsequent steps.
- Place the tip of the ISM in a small, transparent container (either the one the exchanger came in or a small cuvette) holding the liquid ion exchanger (LIX) for TMA. Perform this step under direct visualization using the horizontal microscope setup.
- Apply a small amount of negative pressure to draw a minimal amount of the LIX into the tip (i.e., as soon as LIX is seen entering the tip, stop applying negative pressure).
- Disconnect the 10-mL syringe from the tubing and allow the ISM to sit for 5 min. During that time, the LIX will enter the silanized tip until it reaches a state of equilibrium.
- Remove the ISM from the LIX. Pull the tubing out of the exchanger barrel (while removing as little wax as possible). Place a chloridized silver wire into the small opening created at the back end of the ISM. Seal the wire in the backfill of the exchanger barrel with melted wax.
- Allow the ISM to sit for at least 30 min. Attach completed ISMs to the inside rim of a beaker using any pliable, temporary adhesive.
- Calibrate the ISM by recording the voltage measured by the ISM in each calibration solution made in step 1.8.
NOTE: Calibration can be performed in a calibration station (see step 2.3) or in the experimental setup. This procedure is outlined in Supplement B and in Haack et al10. - If the ISM calibration was successful for multiple ISMs, pause here until the day of intended use. If not, fabricate more ISMs.
- On the day of the experiment, again calibrate the microelectrode (see step 3.29).
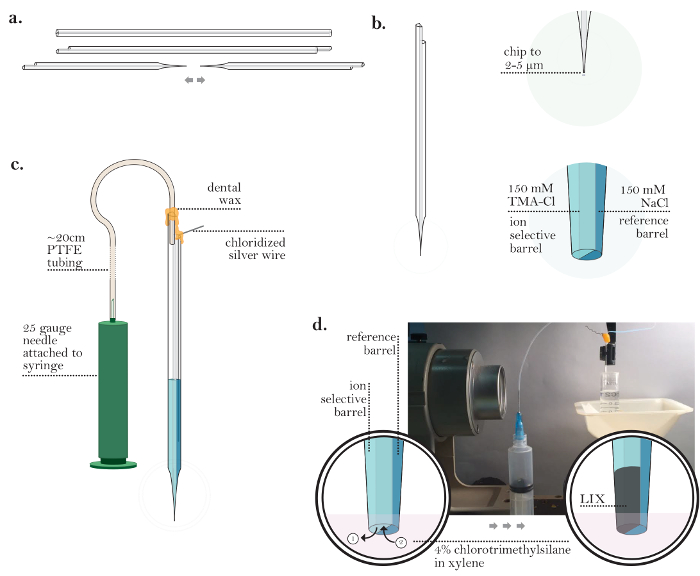
Figure 3: Preparation of an Ion-Selective Microelectrode. (a) ISM after chipping back the ends of a capillary and pulling (steps 3.2-3.6): A single barrel at both ends of a glass capillary is chipped. An ISM is generated by pulling one double-barreled glass capillary to generate two micropipettes with fine tips. (b) ISM after backfilling both barrels (steps 3.7-3.9): The tip of a single ISM is chipped to a diameter of 2-5 µm. The ion-selective barrel is backfilled with TMA-Cl, and the reference barrel is backfilled with NaCl. (c) ISM prior to coating with chlorotrimethylsilane (steps 3.11-3.13): A chloridized silver wire is inserted into the reference barrel. Polytetrafluoroethylene (PTFE) tubing is connected to a 25 G needle and inserted into the ion-selective barrel. An air-tight seal on top of both barrels is created using dental wax. (d) Coating a micropipette with chlorotrimethylsilane (steps 3.15-3.26): [Low magnification] An ISM suspended in chlorotrimethylsilane in line with a horizontally mounted stereomicroscope. [High magnification] The view through a horizontally mounted stereomicroscope of an ISM tip in chlorotrimethylsilane solution. After visualization of the tip through a microscope, small amount of TMA-Cl solution is expelled from the ion-selective barrel (enough to generate a small bubble of TMA-Cl solution). The ISM holder is tapped to release a TMA-Cl solution bubble and then chlorotrimethylsilane is drawn up into the tip. This cycle is repeated several times. After all chlorotrimethylsilane is ejected from the ISM, the ISM is placed into the liquid ion exchanger (LIX) for TMA and LIX is drawn into the tip of the ion-selective barrel. Please click here to view a larger version of this figure.
4. Preparation of Iontophoresis Microelectrodes
NOTE: Iontophoresis microelectrodes should be fabricated on the day of the experiment.
- Pull a double-barreled borosilicate glass capillary on a vertical or horizontal puller. Tailor the parameters to pull a pipette similar to the micropipettes pulled in step 3.6 (Figure 4a).
- Place the micropipette under the compound microscope used in step 3.7 and cut the tip off using a glass microscope slide so that the resulting diameter is between 2 and 5 µm (Figure 4a).
- Fill both barrels with the 150 mM TMA-Cl backfill solution using a 10 mL syringe attached to a 0.22 µm filter and a 28 G, 97 mm needle (Figure 4a).
- Tap the micropipette gently to ensure that no air bubbles are left in the solution of both barrels.
- Place chloridized silver wires into both barrels of the micropipette. Ensure that the wires are deep enough in the backfill solutions so that they will remain in contact with the solutions for the duration of the experiment.
- Seal the wires into the barrels using hot dental wax. Gently interlock the wires by twisting them around each other (completed microelectrode shown in Figure 4b).
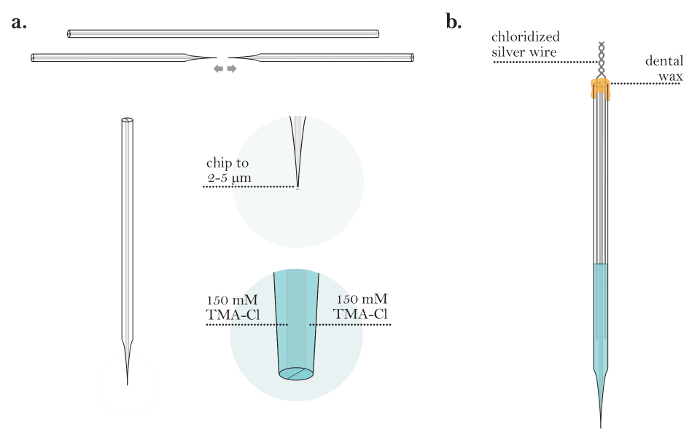
Figure 4: Preparation of an Iontophoresis Microelectrode. (a) Iontophoresis microelectrode after backfilling both barrels (steps 4.1-4.3): An iontophoresis microelectrode is pulled from a capillary tube. The tip of the microelectrode is chipped to a diameter of 2-5 µm. Both barrels of the iontophoresis microelectrode are filled with TMA-Cl solution. (b) Completed iontophoresis microelectrode (steps 4.5-4.6): An iontophoresis microelectrode with two chloridized silver wires inserted into the barrels. The barrels of the microelectrode are sealed with wax, and the silver wires are twisted together at the back of the microelectrode. Please click here to view a larger version of this figure.
5. Preparation of Artificial Cerebrospinal Fluid and Rodent Brain Tissue Slices
- Prepare 1 L of artificial cerebrospinal fluid (ACSF) with a composition appropriate for the experiment and add 0.5 mM TMA-Cl to it.
NOTE: The TMA-Cl is necessary to establish a background concentration of TMA during the experiment. - Prepare rodent brain slices with a thickness of 400 µm according to standard protocols11,12. Use the ACSF prepared in step 5.1 for the dissection and maintenance of brain slices.
6. Real-time Iontophoresis in Agarose
- Turn on the computer running the Walter and Wanda programs.
NOTE: These programs are freely available upon request. While this software is not essential, programming similar software or performing the analysis by hand would otherwise be required. - Run ACSF through the submersion chamber at an appropriate rate (e.g., 2 mL/min). Set the temperature controller to a desired temperature and bubble ACSF with 95% O2/5% CO2 (or another appropriate gas mixture) for the duration of the experiment.
- Mount an indifferent (ground) electrode in a suitable holder and submerge the tip into the ACSF running through the submersion chamber. Connect the wire to the ground of the recording setup.
- Fill the porous cup (made in step 1.7) with the 0.3% agarose prepared previously and place it in the submersion chamber. Ensure that the solution does not run over the top of the cup.
- Secure a calibrated ISM to the pipette holder of one micromanipulator and an iontophoresis microelectrode to the second. Set the holders to an angle appropriate for the setup (Figure 5a).
- Connect the ISM and iontophoresis microelectrode wires to their respective head stages of the recording amplifier. Alternatively, connect directly to the amplifier (depending on the setup).
- Ensure that the weight/positioning of the connecting wires or clips does not cause any movement of the microelectrodes, as small fluctuations in positioning can influence the results.
- Turn on the electronic setup (from step 2). Start Walter and Wanda in separate instances.
- In the Wanda GUI, click "Calibrate" (Figure 6a). In the Calibration box (Figure 6b), fill in the voltages measured during the ISM calibration (step 3.29) and click "Fit Data."
NOTE: This enables fitting to the following representation of the Nicolsky equation (alternatively fit the equation by another means to obtain M and K):
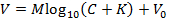
Here, V is the measured voltage (mV), M is the Nicolsky slope (mV), C is the concentration of ion (mM), K is the interference (mM), and V0 is the offset voltage (mV)3. - Click "Accept" in the Calibrate box to automatically transfer the slope (M) and interference (K) generated in step 6.9 to the main GUI.
NOTE: Here, K represents the Na interference, which is usually negligible. - On the left side of the GUI, ensure that all experimental parameters are set in corresponding entries (Figure 6a).
- In the Source Method box, set the source to the iontophoretic source (default), the "Record Duration" to "200 s" (default), the "Pulse Begin" to "10 s" (default), the "Pulse End" to "60 s" (default), the "Bias Current" to "20 nA" (default), the "Main Current" to "100 nA" (default), and the "Conversion Factor" to an appropriate value.
- In the Measuring Electrode box, set "Bath C" to the concentration of TMA contained within the bath solution (expressed in mM). Set the "Total Gain," "Output Channel," "ISM Channel," and "Ref. Channel" to appropriate values for the data acquisition system in use.
NOTE: The "Conversion Factor" must be set to an appropriate value (specific to the iontophoretic unit in use). This value specifies the amount of current that is passed for a given applied voltage from the D/A converter (nA/mV).
- Place a temperature probe in the agar cup. Record the measured temperature in the "Temperature" entry in the "Measuring Electrode" box of the GUI (Figure 6a).
- Turn on the sub-stage illuminator. If required, turn on the camera attached to the microscope and camera monitor.
- Lower the microelectrodes at least 1,000 µm deep into the agarose and center them in the cup (Figure 5b). Visualize them under the microscope using a 10X objective (water-immersion objective with a long working distance).
- Offset the voltage on the amplifier to 0 mV for both reference and ISM channels in order to establish the voltage recorded in the agarose as the baseline voltage.
- On the two-channel amplifier, manually move the ISM channel connector to the voltage subtraction output to set the subtraction 'on' between the reference and ISM channels.
NOTE: Subtraction ensures that the voltage changes in the ISM channel reflect the changes in TMA concentration alone. - Move the ISM so that it touches the tip of the iontophoresis microelectrode. Center the tips on each other in all three directional axes.
- Zero the relative positions of both microelectrodes on the micromanipulator control boxes. Ensure that the microelectrodes are centered accurately and precisely (critical).
- Move the ISM 120 µm away from the iontophoresis microelectrode in one axis (the left-right axis, Figure 5b). Input this distance into the "Measuring Electrode" box of the GUI (Figure 6a).
- Start a recording by clicking "Acquire" in the GUI (Figure 6a); allow the program to record a full recording.
NOTE: The iontophoresis microelectrode receives a constant bias current. After clicking "Acquire," there is a short delay before the main current is applied for a limited duration. - Repeat step 6.20 two to three more times. Wait until the TMA signal returns to baseline before acquiring new records; the program will save each record for later analysis.
- Check the spacing of the two microelectrodes by moving the ISM back to the zero-position specified by the control box. If the microelectrodes are no longer centered, center them again using the same strategy as in step 6.17. Record any changes in the position of the electrodes.
NOTE: If the spacing changes by more than about 2%, the records acquired in step 6.19 cannot be considered accurate and new ones must be taken.
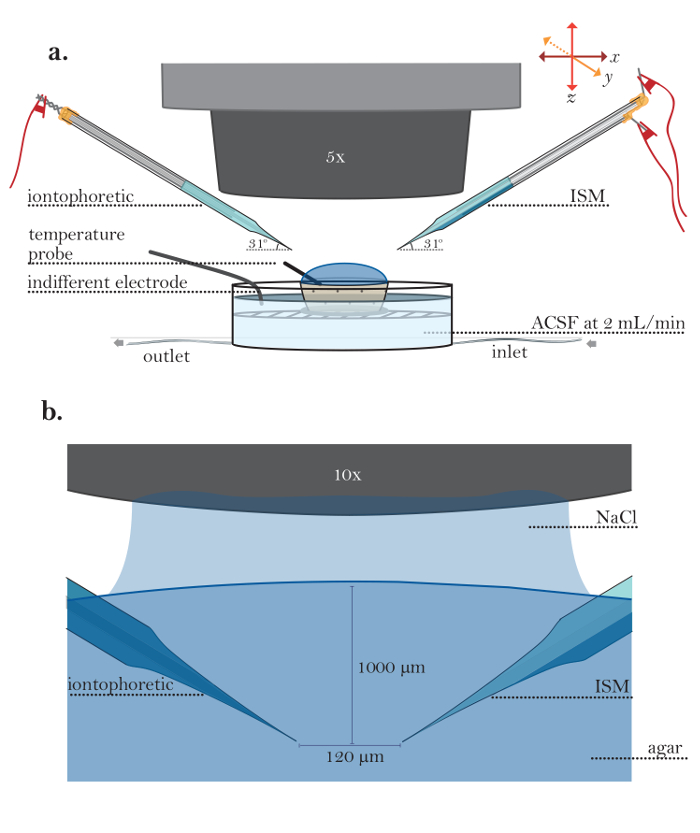
Figure 5: Setup for Experiments in Agar. (a) Setup for experiment in dilute agar (steps 6.1-6.5): A small porous container filled with dilute agar placed in a running perfusion chamber. An iontophoresis microelectrode (left side) and an ISM (right side) are held by microelectrode holders; microelectrode holders are fitted into the arms of robotic micromanipulators. A temperature probe is placed in agar gel, and an indifferent ground electrode is placed within the submersion chamber. (b) Magnified view of microelectrodes in agar: An iontophoresis microelectrode (left side) and an ISM (right side) are visualized in agar using a 10X water immersion objective (objective immersed here in 150 mM NaCl). Microelectrodes are positioned using micromanipulators to a depth of 1,000 µm; the spacing between microelectrodes is 120 µm. Please click here to view a larger version of this figure.
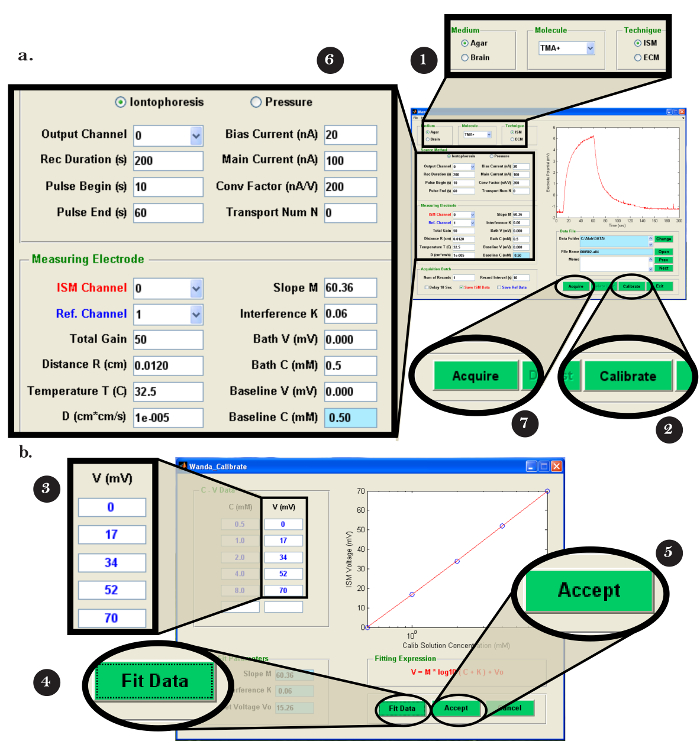
Figure 6: Wanda Computer Software Interface. (a) Navigating Wanda graphical user interface (GUI): The screen that appears after opening the Wanda software. In box (1), the appropriate medium, iontophoresis molecule, and technique are selected. (2) "Calibrate" is clicked to open the Wanda Calibration box. After calibrating the ISM (see Figure 6b and Supplement B), the ISM is positioned in agar or brain, as described in steps 6 and 8 of the protocol. In box (6), all appropriate values for the experiment being performed are entered. (7) "Acquire" is clicked to take a recording; a graph of voltage versus time appears in the top-right portion of the Wanda GUI. (b) Calibrating ISM in Wanda: The window that opens after clicking on (2) "Calibrate" in the Wanda GUI. The values from step 3.29 are entered into box (3), and (4) "Fit Data" is selected. The calibration curve is confirmed to be linear. (5) "Accept" is clicked to return to the Wanda GUI. Please click here to view a larger version of this figure.
7. Agarose Data Analysis
- Open the Walter program on the computer (PC). In the "0. Records From:" menu, click the "Wanda/VOLTORO" button to read the records generated by Wanda (Figure 7a).Assuming that output to a spreadsheet is required, open the appropriate software. Click "Sheet 1,3" in the "1. Write Excel?" menu (Figure 7b).
- In the next pop-up window, select the record(s) to be read and click "Open" (Figure 7b); note that the records will be automatically graphed. To begin the fitting procedure, perform the following steps.
- In the "2. Options" menu, click on the "select rec" button. In the "Figure 2" pop-up window, use the mouse to move the crosshairs over the first record to be processed (Figure 7c); press either mouse button to choose the record.
- Click on "fit curve" in the menu. Select the desired number of iterations of fitting; use at least 20 iterations of fitting to obtain an accurate fit of the data.
- In the menu, select "all" to fit all data points and select "continue;" the program will fit the displayed curve. Observe the fitting procedure and compare the experimental record with the best fitted curve obtained.
- Select the option to write the result to the appropriate spreadsheet program by clicking "Excel" in the "7. Results" menu (Figure 7d). Note (and record) the following critical data that will be used to determine the functionality of the iontophoresis microelectrode: 'D(E5)', 'Reference D(E5)', 'r_app', transport number 'nt', 'Apparent nt'.
NOTE: "D(E5)": Measured free diffusion coefficient x 105 (cm2/s); "Reference D(E5)": Theoretical free diffusion coefficient x 105 (cm2/s). This value is extracted from a database within Walter based on the ion, the medium, and the temperature input. "r_app": Apparent microelectrode spacing (cm), calculated based on the measured and reference D(E5). "nt": Transport number (dimensionless). This number determines the fraction of the iontophoresis current that is being used to release TMA4. "Apparent nt": Apparent transport number (dimensionless). This is a transport number calculated from r_app. This number should be close to the measured nt. - Repeat steps 7.1-7.3 for each of the records for a chosen pair of microelectrodes.
- Determine whether the iontophoresis microelectrode is usable by doing the following.
- Compare "r_app" with the actual r (i.e., 120 µm); this criterion is fulfilled if the average values from all trials are within 4% of each other.
- Compare "D(E5)" with the Reference D(E5); this criterion is fulfilled if average values from all trials are within 8% of each other.
- Compare the "nt" between trials with the same microelectrode; this criterion is fulfilled if average values from all trials are within 10% of each other.
- If one of the criteria from step 7.5 was not fulfilled, troubleshoot the iontophoresis microelectrode or begin testing another one.
- If the iontophoresis microelectrode is deemed suitable for the experiment, record the average transport number from all trials in the "Transport Num N" field in the Wanda GUI (Figure 6a).
Figure 7: Walter Computer Software Interface. (a) Choosing the data collection program in Walter: The "0. Records From:" menu opens after starting the Walter software. The option to load the records saved by Wanda is selected by clicking the "Wanda/Voltoro" button. (b) Choosing the data and data analysis output location in Walter: [Left] After the appropriate spreadsheet program is opened, "Sheets 1,3" is chosen to output all Walter data analysis to the previously opened spreadsheet program. [Right] After the data analysis output location is chosen, a pop-up window opens, allowing the user to choose the first and last recordings to be read by Walter. (c) Choosing the recording to analyze in Walter: [Right] After the files to read are chosen, a pop-up window will open with all chosen records displayed as a graph ("Figure 2"). [Left] In the "2.Options" menu, "select rec" is clicked, and the mouse is used to move the crosshairs to identify the first recording for analysis; either mouse button is pressed to choose the recording. (d) Exporting the data analysis from Walter to a spreadsheet: After fitting the data, a pop-up window and the "7. Results" menu appear. [Left] Graph of the selected recording (blue) with the fitted diffusion curve generated by Walter (red). [Right] The "7. Results" menu allows the user to write the data from the analysis to a spreadsheet program by clicking the "Excel" button. Please click here to view a larger version of this figure.
8. Real-time Iontophoresis in Brain Slices
- Place a 400 µm-thick brain slice in the recording chamber, ensuring that it is fully submerged in the flowing ACSF. Position the slice using a watercolor paintbrush and gently secure it with a grid.
- Move both the iontophoresis microelectrode and the ISM above the field of interest on the brain slice. Submerge both in the flowing ACSF but above the slice.
- Offset the voltage for both the reference and the ion-sensing channels to "0" mV. Wait for the voltage in both channels to stabilize. On the chart recorder, mark the voltage measured on the ion sensing channel of the ISM. Use this to calculate the baseline V parameter in Wanda.
- Place the ISM and iontophoresis microelectrode 200 µm deep in the slice and 120 µm away from each other. Wait for the stabilization of the signal after moving the microelectrode into the brain slice.
NOTE: The bias current applied to the iontophoresis microelectrode causes a small accumulation of TMA. It is a common mistake to take a recording too soon and underestimate the signal buildup. - On the chart recorder, mark the stabilized voltage measured in the brain slice on the ion-sensing channel of the ISM. Calculate the voltage difference between the TMA signal measured in step 8.3 and step 8.4 and input this value into the "Baseline V (mV)" field in the Measuring Electrode box of the Wanda GUI (Figure 6a).
- On the left side of the GUI, ensure that all experimental parameters are correctly recorded/entered. Set "Medium" to "Brain," "Transport number" to the average value calculated for the iontophoresis microelectrode in step 7.4, and "Temperature" to the temperature of the bath containing the slice.
NOTE: V must be recorded for each set of measurements. The baseline V will be converted by Wanda into the baseline C (mM) parameter (i.e., the concentration of TMA in the brain tissue). - Start the recording by clicking "Acquire" and allow it to take a full recording. Wait until the TMA signal returns to baseline before acquiring a new recording.
- Take two to three successive recordings before removing the microelectrodes from the chosen brain location. Input the temperature measured into the Wanda software immediately before each recording.
- Move both microelectrodes diagonally back to the surface of the slice. Raise both to at least 50 µm above the slice. Using the chart recorder, determine any change between the V measured now and its measurement from step 8.3.
- Center the tips of the ISM and the iontophoretic microelectrodes relative each other in the x-, y-, and z-axes. Obtain spacing changes, if any, from the display of the micromanipulator control box.
9. Brain Data Analysis
- Open a new spreadsheet for the analysis output.
- Repeat steps 7.1-7.4 in Walter to analyze the recordings taken from the brain.
- Write the data onto the spreadsheet program by clicking "Excel" in the Walter menu. Record the α, volume fraction of the brain ECS; λ, tortuosity of the brain ECS; and k (s-1), non-specific clearance.
10. Checking Transport Number and ISM Calibration
- Measure the ISM transport number (nt) at the end of the experiment using the protocol below. Alternatively, check nt after critical trials or when measurements appear anomalous. However, checking nt too many times can result in trauma to the brain slice.
- Take new recordings in agarose. See steps 6.4, 6.11, 6.12, 6.14, 6.15, and 6.17-6.22.
- Repeat steps 7.1-7.4 in Walter to obtain the nt from the new agarose recordings. Inspect the spreadsheet: if the nt has changed by more than 10% from the nt obtained prior to the brain measurements, the data obtained with this iontophoretic microelectrode are not reliable.
- Perform a new calibration (see step 3.29) for the ISM after all brain data has been collected. Use newly obtained ISM calibration data as the input in the Wanda Calibrate box (see steps 6.9 and 6.10) and check that the slope value differs by less than 10% from the previous calibration.
NOTE: The data obtained with this ISM are not reliable if the slope value differs by more than 10% from the previous calibration.
Representative Results
The utility of the RTI technique is demonstrated in an experiment designed to measure the changes in α and during a hypoosmolar challenge (Figure 8 and Figure 9). It has previously been shown that reducing the osmolarity of the ECS by washing on hypotonic ACSF will produce a decrease in α and an increase in λ13.
In this experiment, RTI was performed on rat brain slices under both control conditions and during the wash-on of hypotonic ACSF. An ISM was fabricated, and its calibration parameters were input into Wanda for fitting to the Nicolsky equation, which calculated a slope (M) of 58.21 mV. The ISM and iontophoresis microelectrodes were placed in agar and positioned 120 µm apart from each other in order to measure the transport number. Three recordings were taken, and the curves were fitted and analyzed according to the procedure in step 6 of the protocol (Figure 8a). The fitted curve of each trial overlapped with the raw curve (Figure 8a). The measured diffusion coefficient (D x 1E5), the transport number (nt), and the difference between the apparent spacing of the microelectrodes (r_app) and their actual spacing (r) did not differ significantly between the three recordings (Figure 8b, recordings a1-3). Based on these criteria, this iontophoresis microelectrode was deemed acceptable to continue with the experiment.
Once the stable iontophoresis microelectrode was chosen, control values for α and λ in the rat brain slice were taken in order to establish a baseline for these parameters. Previous studies found control values for the rat neocortex to be α = 0.18-0.22 and λ = 1.54-1.651. To replicate these values in this experiment, the ISM and iontophoresis microelectrode were placed 200 µm deep in the rat neocortex and 120 µm apart from each other. The average nt, calculated from the data in Figure 8b, was entered into the Wanda program for use in the calculations of α and λ. A shift in baseline V from the placement of the two microelectrodes about 200 µm deep in the brain was recorded, and the voltage jump was entered into Wanda to correct the baseline TMA (i.e., the baseline C parameter) concentration. Three recordings were taken, and their curves were fitted (Figure 9a, Figure 9d, and Figure 9f). The fits revealed an average α = 0.192 and λ = 1.69 (Figure 9e). Spacing and shifts in baseline V were checked after the recordings were taken, and the corrected values were entered into Wanda to reanalyze the data (as detailed in step 8 of the protocol). The recalculated values did not differ significantly, and the values reported in Figure 9d were accepted.
The normal osmolarity of ACSF is 300 mOsm. To test the effect of hypotonic ACSF on α and λ in the rat somatosensory neocortex, ACSF with an osmolarity of 150 mOsm was made by reducing the NaCl concentration. It was hypothesized that this hypotonic ACSF would lead to swelling of brain cells, causing a lower α and a potentially higher λ13. The brain slice was superfused with hypotonic ACSF for approximately 30 min, allowing it to equilibrate with the brain. During this time, the microelectrodes remained in the same place in the neocortex as they were during previous measurements of control conditions. Five recordings were taken under hypotonic conditions (Figure 9b and f). This generated an average α = 0.13 and λ = 1.84 (Figure 9e). These values were consistent with the hypothesis that hypoosmolarity decreases α and increases λ. Spacing and changes in baseline V were measured and taken into account during the analysis and fitting procedure.
Recovery parameters were also measured by washing on regular ACSF (300 mOsm) and taking new recordings in the same place in the neocortex. Because swelling effects should be reversible, it was expected that α and λ would recover to control levels. The values averaged over four records taken after 30 min of regular ACSF wash-on were α = 0.37 and λ = 1.61 (Figure 9c, Figure 9e, and Figure 9f). This demonstrated that there was an unexpected overshoot during the recovery of α under these conditions (Figure 9e and Figure 9f). Afterwards, the microelectrodes were returned to agar to confirm that the transport number of the iontophoresis microelectrode was unchanged (Figure 8c). The ISM was then recalibrated, and the new fit to the Nicolsky equation revealed the slope to be 58.21 mV.
This experiment is a clear example of what RTI looks like under ideal conditions. The following elements of the experiment were the key to its success. First, experimental data collected in agarose and the brain demonstrated adequate overlap with the theoretical curves generated by Wanda (Figure 8a and Figure 9a and Figure 9c). The similarity in slope, peak, and return to a similar baseline are all important in determining the strength of the match. These portions of the curve are frequently problematic when recording in agarose, and it is common that multiple recordings must be taken before finding the conditions that produce well-matched curves (i.e., good microelectrodes). Second, the average transport numbers before and after the experiment were within 10% of each other (Figure 8b and Figure 8c). If this had not occurred, the values recorded in brain could not be trusted. This is by far the most common problem that occurs in RTI experiments. Third, the ISM calibrations in standardized TMA solutions before and after the experiment matched (data not shown). Typically, the calibrations of a working ISM are within 10%, making this an uncommon source of experiment failure.
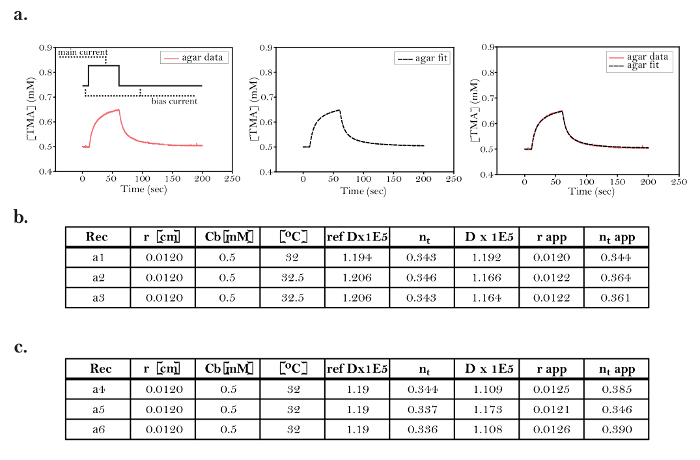
Figure 8: Ideal Curve Fitting Data in Agar Before and After Experimentation in the Brain. (a) Representative data from a trial in agar: [Far left] Representative data from a single trial obtained in agar demonstrating the concentration curve of TMA. Prior to diffusion measurements, a constant bias current of +20 nA was applied through the iontophoresis microelectrode. At time = 10 s, TMA was pulsed from the iontophoresis microelectrode into the agar by applying a +60 nA main current for 50 s. A diffusion curve was generated by measuring [TMA] over time using an ISM positioned 120 µm from the source. [Middle] A fitted curve obtained from data processing in Walter. [Right] The overlap of the data and the fitted curve demonstrates that the curve fitting done by Walter accurately models diffusion in this trial. (b) Table of agar measurements before experimentation in the brain: Data obtained from three trials (a1, graphed above) prior to the hypotonic stress experiments (Figure 9). All trials were conducted with the iontophoresis microelectrode and ISM used for the hypoosmotic stress trials. The data fulfilled the criteria needed to proceed with the experiment in brain slices. These criteria include adequate overlap between the data and the fitted curve (as above) and less than 10% variation in transport number. Additional criteria are outlined in step 7.6. (c) Table of agar measurements after experimentation in the brain: Data obtained from three trials performed in agar after the hypoosmotic stress experiments (Figure 9). The consistency demonstrated between trials a1-3 and a4-6 strongly suggests that the ISM and iontophoresis microelectrodes were stable throughout the brain trials. Rec = recording or trial; r = distance between the ISM and iontophoresis microelectrode; Cb = baseline concentration; ref Dx1E5 = theoretical free diffusion coefficient x 105 (cm2s-1) based on a pre-calculated standard; nt = transport number (dimensionless); D(E5) = measured free diffusion coefficient x 105 (cm2s-1); r_app = apparent microelectrode spacing (cm) based on the measured and reference D(E5); nt apparent = apparent transport number based on r_app. Please click here to view a larger version of this figure.
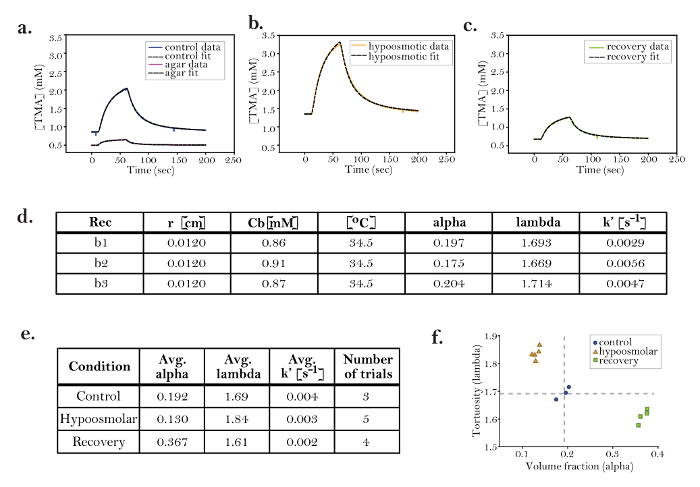
Figure 9: Hypoosmotic Stress Decreases Alpha and Increases Lambda
a-c. Representative data from trials in the brain under (a) control, (b) hypoosmotic, and (c) recovery conditions: Solid lines represent data and dashed black lines are fitted curves. The three conditions demonstrate markedly different diffusion curves, including different slopes, amplitudes, and widths. (d) Data table from control trials: Data table of three control trials (b1, graphed above); α and λ are similar in all trials and consistent with published data for the rat neocortex. For all trials in the brain, the average nt from pre- and post-experiment agar measurements (Figure 8b and 8c) was used for the brain nt. The brain Dref was set to 1.25 × 10-5 cm2 s-1, based on a database of diffusion coefficients (in Walter) that was obtained in the rat brain when T = 34.5 [°C]. The parameter k' [s-1] accounts for the small amount of TMA lost from the ECS during the diffusion measurements. Although k' is typically very small, including the parameter in curve fitting improves the accuracy of the RTI method. The loss parameter k' probably represents cellular uptake or the loss of TMA to the ACSF. (e) Comparison of control, hypoosmotic, and recovery conditions: Averages from all trials in the brain under control, hypoosmotic, and recovery conditions. The data demonstrate that hypoosmotic stress decreases α and increases λ. During a recovery period following hypoosmotic conditions, α overshoots baseline (control), while λ returns to baseline. The results suggest that changes in ECS during hypoosmotic challenges are partially reversible. The RTI method is ideal for studying this type of acute reversible effect. (f) Graph demonstrating data clustering: The volume fraction (x-axis) and tortuosity (y-axis) from each trial are plotted as a single point. The graph demonstrates the clustering of data within each group (i.e., control, hypoosmolar, and recovery), suggesting that RTI has the sensitivity to detect the reproducible effects of a hypoosmolar challenge in the brain ECS. Rec = recording or trial; r = distance between ISM and iontophoresis microelectrode; Cb = baseline concentration; alpha = volume fraction; lambda = tortuosity; k' = non-specific clearance of probe. Please click here to view a larger version of this figure.
Supplemental Files: Please click here to download the files.
Discussion
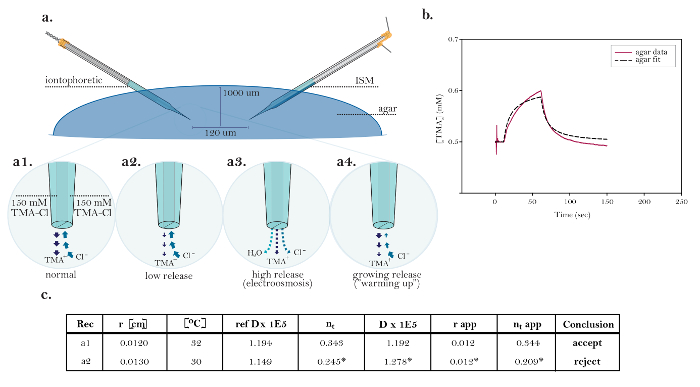
Figure 10: Non-ideal Data Demonstrating Common Technical Issues. (a) Diagrams of common technical issues with iontophoresis microelectrodes: Comparison of the normal release of TMA from a functioning iontophoresis microelectrode with three sources demonstrating technical issues. [High magnification, a1] The current in an ideal iontophoretic source is carried equally by TMA release and chloride uptake. [High magnification, a2] An iontophoresis microelectrode with low nt releases less TMA and takes up more chloride than normal. [High magnification, a3] An iontophoresis microelectrode displaying electroosmosis releases TMA, chloride, and solvent. [High magnification, a4] An iontophoresis microelectrode displaying growing release over time (i.e., "warming up"). (b) Graph of non-ideal data obtained in agar: The data is not adequately modeled by the curve fitted by Walter and therefore cannot be accurately interpreted; the exact cause of the discrepancy is unclear. (c) Table of non-ideal data obtained in agar: Normal or expected results in agar are displayed in the top row (graphed in Figure 8a) for comparison to the non-ideal data in the second row (graphed in Figure 10b). The poor overlap between the data and the fitted curve in Figure 10b means that the fitted curve does not accurately model the diffusion data; therefore, the calculated values (marked with *) cannot be interpreted. This could have been caused by issues with the iontophoresis microelectrode (e.g., warming up) or the ISM (e.g., slow response). Troubleshooting: exchange microelectrodes one at a time, starting with the iontophoresis microelectrode. Rec = recording or trial; r = distance between ISM and iontophoresis microelectrode; Cb = baseline concentration; ref Dx1E5 = theoretical free diffusion coefficient x 105 (cm2s-1) based on a pre-calculated standard; nt = transport number (dimensionless); D(E5) = measured free diffusion coefficient x 105 (cm2s-1); r_app = apparent microelectrode spacing (cm) based on the measured and reference D(E5); nt apparent = apparent transport number based on r_app. Please click here to view a larger version of this figure.
While the experiment shown in Figure 8 and Figure 9 had a stable and working iontophoresis microelectrode and ISM, there are many experiments in which either or both microelectrodes are compromised and do not yield ideal results. A "normal" TMA iontophoresis microelectrode has a value of nt  0.3. Figure 10a demonstrates three common issues with the iontophoresis microelectrode that can be encountered during RTI experiments.
0.3. Figure 10a demonstrates three common issues with the iontophoresis microelectrode that can be encountered during RTI experiments.
Low release. The iontophoresis microelectrode releases very little TMA when bias current or main current is applied, resulting in nt <0.1. Current is still passing through the tip, but most of it is carried by the Cl anion entering the tip and very little by the TMA cation leaving the tip. If nt is stable in several consecutive trials, these iontophoresis microelectrodes can be used. However, this is not recommended, as they are not functioning optimally, meaning that further issues may develop. An even bigger extreme occurs when the tip of the iontophoretic microelectrode is blocked and no ions leave or enter the tip. In this case, no curve will be produced. In such instances, after checking that all electrical connections are proper and secure, the iontophoresis microelectrode should be discarded.
High release (electroosmosis). In addition to TMA, the iontophoresis microelectrode also releases water, resulting in nt > 0.5. If the nt is stable over several trials, these iontophoresis microelectrodes can be used, but this is not recommended, as further issues may develop. The only troubleshooting step to take is to reduce the main current. This sometimes eliminates water release and causes the nt to decrease below 0.5.
Growing release ("warming up"). In this case, the TMA release increases over time. When the "warming up" is rapid, the diffusion curve has a shape similar to that shown in Figure 10b, and it cannot be reliably fitted. In this case, the diffusion curve demonstrates a slow rise in TMA concentration during the initial phase of the main current, and the TMA concentration does not plateau. An unreliable fit creates an inaccurate measured D, which affects the consistency of the measured transport number and the r_app values. When the "warming up" is more gradual, it does not have a significant impact on the shape of individual diffusion curves, but it manifests in an nt that increases over successive trials. A "warming up" condition can sometimes be remedied by "pulsing" the iontophoresis microelectrode for a period of time (about 30 min). This is done by alternating between a bias current and a high main current (+200 nA) for a few seconds at a time. If an iontophoresis microelectrode still does not give a stable transport number, it is best to simply test a new one.
Precise measurement of the transport number and stability during the entire experiment are essential to ensure an accurate value for α. Maintaining the spacing between microelectrodes is critical to the determination of both α and λ. If the spacing does change after a measurement, either in agarose or in the brain, the straight-line distance between the tips of the microelectrodes can be entered into the output spreadsheet and reanalyzed by Walter. If the values differ too much, the measurement must be discarded. Temperature fluctuation can also be a contributing factor to inaccuracy, so using an accurate temperature probe and a reliable chamber heating element is important.
The iontophoresis microelectrode is the most frequent source of problems in the RTI technique; making and using a stable ISM is crucial to obtaining good data. One possible problem with the ISM can be a sluggish response, which can be caused by very high impedance in the tip. With a slow-responding ISM, all the iontophoresis microelectrodes will appear to have a "warming up" effect (Figure 10b), but the curve is simply caused by the inability of the ISM to detect the changing TMA concentrations fast enough. Increasing the distance between the microelectrodes (up to 150 µm) can allow more time for the ISM to respond and can improve curve fitting. A sluggish response may indicate that the ion exchanger has retreated up inside the tip. This can be seen under a compound microscope and, if present, means the silanization was poor and that the ISM must be discarded. In addition, drifts in the ISM signal can cause inaccurate fits of the data. It is up to the experimenter to determine if the drift is affecting the data beyond tolerance.
Limitations of RTI
There are several limitations to the RTI method because of assumptions underlying the data analysis. These assumptions include a requirement for tissue homogeneity and tissue isotropy in both the brain region of interest and a spherical volume surrounding this region. In the context of RTI, tissue homogeneity requires that the diffusion parameters are constant within the region of interest. Tissue isotropy means that a single value of D* applies to all three spatial axes. Each molecule released from a source microelectrode takes a random path before arriving at the position of the recording ISM. The voltage on the ISM, representing the number of molecules (i.e., concentration) recorded at a single time, includes molecules that have traveled in all three spatial axes, as well as some molecules that have traveled beyond the ISM and have returned to the measuring point (Figure 1c). During RTI data analysis, the Walter program generates average α and λ, which include the diffusion of all molecules traveling in all axes from a point source to the ISM. If the rate of diffusion is significantly different in any one of the three spatial axes (anisotropy) or if the tissue is non-homogeneous, additional data collection and data analysis are required to calculate α and λ8,14.
In addition to the above tissue prerequisites, the RTI method requires that the spacing between a point source and ISM, which is referred to as r, is roughly 80 – 130 µm. When r is decreased below 50 µm, the ISM response may not be fast enough to record diffusion-dependent changes in concentration of the probe molecule. This might be remedied in the future using concentric ISMs with faster response times10,15. Larger r distances also minimize brain region-independent differences in the ECS milieu, ISM tip size, and brain tissue damage during ISM placement. Conversely, when r is increased beyond 150 µm, the diffusion of molecules from the iontophoretic point source is more susceptible to influence by non-isotropic, inhomogeneous elements surrounding the brain region of interest or the tissue-perfusate boundary14.
Incorporating RTI and alternative techniques to explore ECS
The RTI method belongs to a larger group of techniques that utilize a molecular probe to study the ECS; each method has its own advantages and shortcomings. While RTI allows for an accurate calculation of both α and λ in real time, the method requires a charged molecular probe that can be detected by an ion exchanger. In experiments where iontophoresis is not suitable, such as the study of an uncharged probe, iontophoresis may be replaced by pressure ejection. Unfortunately, current techniques do not allow for the calculation of α with pressure ejection, because the volume released depends on the properties of the injected medium16. To use a probe for which no exchanger exists, the probe may be fluorescently tagged and its diffusion through ECS measured by epifluorescent microscopy. This technique, known as integrative optical imaging (IOI), is limited by the size and availability of fluorescently labeled molecules and the potential for cellular uptake17,18. The IOI technique has the advantage that macromolecules can be used as probes, and this has revealed that λ increases with molecular size. Finally, an important class of diffusion methods has employed radiotracers, but they are no longer in common use2.
Future Applications of RTI
The RTI method can be difficult to implement, and it demands persistence, but it is a powerful tool for quantifying changes that occur in the parameters that describe the brain ECS. This protocol describes the RTI method as applied to slices, but it is also possible to reliably implement this technique in vivo, expanding its potential1,4,6. It can also be used to test the effects of a wide variety of changes in brain physiology, such as those induced by alterations to the chemical environment, pharmacology, trauma, or genetic knockout1. As long as the change induced in the ECS lasts for a period of about 2 min or more, RTI can provide a precise quantification of ECS volume fraction and tortuosity.
While significant insights into the structure and function of brain ECS have been generated in the last 50 years, there remain many unanswered questions. For example, it is still unclear if and how homeostatic mechanisms regulate α and how changes in α affect brain function. Computer models have helped to estimate the relative contributions of cell geometry and other factors that influence λ, but more work is needed1. Finally, the role of the ECS in the pathogenesis of neurological disease (and vice versa) is largely unexplored. In the near future, RTI measurements might improve targeted drug delivery to specific brain regions19.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The work was supported by NIH NINDS grant R01 NS047557.
Materials
| A/D and D/A converter | National Instruments Corporation | NI USB-6221 DAQ | The NI USB-6221 is still sold as a 'Legacy' device by NI. They recommend using NI USB-6341 X Series DAQs for new installations, however we have not tested the newer units. We describe the use of the NI USB-6221 with MATLAB and Windows 7 (32-bit). Alternatives: the much older PCI-MIO-16E-4 A/D converter (Used under Windows XP or older OS only) with BNC-2090 BNC connector panel and SH68-68-EP cable. As noted in the Wanda Manual, an experimental MATLAB program to use Axon Binary Files is available. |
| agarose | Lonza | NuSieve GTG Agarose #50081 | to prepare dilute agarose gel for RTI measurements |
| amplifier for ISM | Dagan | Model IX2-700 Dual Intracellular Preamplifier | ion and reference voltage amplifier with N=0.1 (for reference barrel) and N=0.001 (for ion barrel) headstages |
| biological compound miscroscope (with 4x and 10x objective) | for chipping the microelectrode tips and inspecting microelectrodes; various suppliers, e.g. AmScope | ||
| borosilicate theta capillary glass tubing | Harvard Apparatus | Warner Instruments model TG200-4; order #64-0811 | double-barreled glass tubing for ion-selective microelectrodes and iontophoretic microelectrodes; O.D. 2.0 mm, I.D. 1.4 mm, septum 0.2 mm, length 10 cm |
| brush | Winsor & Newton | University Series 233, size 0 | round shoft handle brush, available from Amazon |
| bunsen burner | Fisher | ||
| camera for visualizing micropipettes | Olympus | OLY-150 | requires monitor, IR filter on substage illuminator is optional |
| chart recorder | to record continuously voltages on ion-selective microelectrode during calibration in tetramethylammonium standards and during RTI experiment; e.g. Kipp & Zonen type BD112 dual-cannel chart recorded, available refurbished | ||
| chlorotrimethylsilane, puriss., > 99% | Sigma-Aldrich | catalog # 92360 | for silanization; CAUTION: flammable, acute toxicity (oral, dermal, inhalation), skin corrosion, eye damage, reacts violently with water, see Sigma-Aldrich Safety Information for full description |
| Commercial Software | The MathWorks | MATLAB, Data acquisition toolbox | for data acquisition and analysis using Wanda and Walter programs. Note that an academic license is available. |
| eye protective goggles | Fisher | ||
| fixed-stage compound microscope | Olympus | BX51WI | can use other compound microscopes with fixed stages |
| forceps | Fine Science Tools | #11251-10 | to chip glass capillary; Dumond #5, preferably used and no longer needed for fine work |
| fume hood | for silanization and filling the tip of ion-selective barrel with liquid ion exchanger; various supliers, e.g. Captair with approriate filter sold by Erlab | ||
| glass microscope slide | Fisher | #12-550A | to chip microelectrode tips |
| heater/stirrer | Fisher | Corning PC-420D | to prepare dilute agarose gel and stir solutions |
| iontophoretic unit | Dagan | ION-100 and PS-100 | ION-100 is a single channel iontophoresis unit +/- 130 V compliance; PS-100 is an external power supply; alternatives: e.g. Axoprobe-1A made by Axon Instruments (now Molecular Devices), out of production, check for availability of refurbished units (eBay and other sites) |
| liquid ion exchanger (LIX) for tetramethylammonium | World Precision Instruments | IE190 Potassium Ion Exchanger | Note: this is equivalent to the original Corning potassium exchanger 477317 based on tetraphenlyborate – do not confuse with neutral carrier potassium exchanger originating from the laboartory of Dr. Simon, ETH, Zurich, which does not sense tetramethylammonium, and is sold by Fluka. You can also make liquid ion exchanger for tetramethylammonium yourself: 3% by weight potassium tetrakis = (p-chlorophenyl) borate dissolved in 2,3-dimethylnitrobenzene. Buy chemicals from Fluka (now part of Sigma). See Oehme and Simon (1976) Anal. Chim. Acta 86: 21-25; CAUTION: The toxicological properties of this liquid ion exchanger have not been fully determined. Ingestion or contact with the human body may be harmful. Exercise due care! Liquid ion exchangers should be stored in a cool place out of direct sunlight. |
| microelectrode holder | WPI | M3301EH | to hold ion-selective microeletrode prefabricate for silanization and filling the tip of ion-selective barrel with liquid ion exchanger; WPI sells two versions of this holder, clear M3301EH and black M3301EH. In our experience, the clear M3301EH appears to be sturdier then the black M3301EH. |
| micromanipulator | Narishige | MM-3 | to position ion-selective microelectrode prefabricate during silanization and filling the tip of ion-selective barrel with liquid ion exchanger; can be substituted with any three-axis micromanipulator in good working condition |
| micropipette puller | Sutter Instruments | Model P-97 | to pull double-barreled glass tubing; other pullers can be used as long as they can accommodate large diameter double-barreled glass tubing |
| microprobe thermometer | Physiotemp | Model BAT-12R | fine probe of this thermometer is placed close to recording site |
| needle | BD | Syringes and Needles # 305122 (25 gauge) | for silanization; BD PrecisionGlide needles 25 G x 5/8 in (0.5mm x 16mm) |
| objective 5x dry | Olympus | MPlan N | |
| objective 10x water immersion | Olympus | UMPlan FL N | 10x objective is water immersion, numerical aperture is 0.3, working distance is 3.3 mm |
| plastic containers (with lids) | Fisher | #14-375-148 | to store tetramethylammonium standard solutions and microelectrodes |
| platform and x-y translation stage for fixed-stage microscope | EXFO | Gibraltar Burleigh | platform holds slice chamber, micromanipulators and accesorries, x-y translational stage moves microscope without compromising recording stability |
| porous minicup | for RTI measurements in a dilute agarose gel; homemade | ||
| reusable adhesive | Bostik | Blu-Tack | for securing microelectrodes to holding vessel and other uses; various suppliers, available from Amazon |
| robotic micromanipulator with precise x,y,z positioning | Sutter Instruments | MP-285 | two mircomanipulators are needed to hold separately ion-selective microelectrode and iontophoretic microelectrode. Also possible to glue micropipettes in a spaced array (see text). |
| signal conditioning unit with low-pass filter | Axon Instruments | CyberAmp 320 or 380 | no longer available from the manufacturer but may be available from E-Bay; alternatives: e.g. FLA-01 Filter/Amplifier from Cygnus Technology. This is a single channel instrument with a minimum cutoff at 10 Hz using a multipole Bessel filter but the company may be willing to modify it for a lower cutoff frequency (2 Hz) if needed. |
| silver wire | A-M Systems | #7830 | diameter 0.015", bare (no coating) |
| slice chamber | Harvard Apparatus | Warner Model RC-27L | this is submersion slice chamber; do not use interface slice chamber |
| stereomicroscope | for silanization and filling the tip of ion-selective barrel with liquid ion exchanger; horizontally mounted; various suppliers | ||
| syringe, 10 mL | BD | Syringes and Needles #309604 | to backfill microelectrodes and for silanization; BD Luer-Lok tip |
| syringe filter 0.22µm pore | Whatman | #6780-1302 | to filter backfill solutions; available from Fisher |
| syringe needle, 28 gauge, 97mm | World Precision Instruments | MicroFil MF28G-5 | to backfill microelectrodes |
| Teflon (=PTFE) tubing | Component Supply | STT-28 PTFE tube light wall (28 gauge) | for silanization of ion-selective barrel; fits on BD PrecisionGlide needles 25 G x 5/8 in. Note: Teflon is essential, PVC tubing would melt by hot wax. |
| temperature control system | Harvard Apparatus | Warner Models TC-344B and SH-27A | TC-344B is a dual automatic temperature controller, SH-27A is an in-line heater; controller and heater work with Warner slice chambers |
| tetramethyammonium (TMA) chloride | Sigma-Aldrich | T-3411 | 5 M solution; CAUTION: acute toxicity (oral, dermal, inhalation), carcinogenicity, hazardous to the aquatic environment, see Sigma-Aldrich Safety Information for full description |
| vibrating blade microtome | Leica | VT1000S | to cut brain slices |
| xylenes | Fisher | X5-1 | for silanization; CAUTION: flammable, acute toxicity (oral, dermal, inhalation), skin corrosion, eye damage, carcinogenicity, see Fisher Safety Information for full description |
References
- Sykova, E., Nicholson, C. Diffusion in brain extracellular space. Physiol Rev. 88 (4), 1277-1340 (2008).
- Nicholson, C. Diffusion and related transport mechanisms in brain tissue. Rep Prog Phys. 64 (7), 815-884 (2001).
- Nicholson, C. Ion-selective microelectrodes and diffusion measurements as tools to explore the brain cell microenvironment. J Neurosci Methods. 48 (3), 199-213 (1993).
- Nicholson, C., Phillips, J. M. Ion diffusion modified by tortuosity and volume fraction in the extracellular microenvironment of the rat cerebellum. J Physiol. 321, 225-257 (1981).
- Nicholson, C., Sykova, E. Extracellular space structure revealed by diffusion analysis. Trends Neurosci. 21 (5), 207-215 (1998).
- Xie, L. L., et al. Sleep drives metabolite clearance from the adult brain. Science. 342 (6156), 373-377 (2013).
- Hrabetova, S., Nicholson, C., Michael, A. C., Borland, L. M. Biophysical properties of brain extracellular space explored with ion-selective microelectrodes, integrative optical imaging and related techniques. Electrochemical Methods for Neuroscience Neuroscience. , 167-204 (2007).
- Rice, M. E., Okada, Y. C., Nicholson, C. Anisotropic and heterogeneous diffusion in the turtle cerebellum: implications for volume transmission. J Neurophysiol. 70 (5), 2035-2044 (1993).
- Vargova, L., et al. Diffusion parameters of the extracellular space in human gliomas. Glia. 42 (1), 77-88 (2003).
- Haack, N., Durry, S., Kafitz, K. W., Chesler, M., Rose, C. Double-barreled and concentric microelectrodes for measurement of extracellular ion signals in brain tissue. J Vis Exp. (103), (2015).
- Xiao, F., Hrabetova, S. Enlarged extracellular space of aquaporin-4-deficient mice does not enhance diffusion of Alexa Fluor 488 or dextran polymers. Neuroscience. 161 (1), 39-45 (2009).
- Sherpa, A. D., Pvan de Nes, ., Xiao, F., Weedon, J., Hrabetova, S. Gliotoxin-induced swelling of astrocytes hinders diffusion in brain extracellular space via formation of dead-space microdomains. Glia. 62 (7), 1053-1065 (2014).
- Kume-Kick, J., et al. Independence of extracellular tortuosity and volume fraction during osmotic challenge in rat neocortex. J Physiol. 542 (Pt 2), 515-527 (2002).
- Saghyan, A., Lewis, D. P., Hrabe, J., Hrabetova, S. Extracellular diffusion in laminar brain structures exemplified by hippocampus. J Neurosci Methods. 205 (1), 110-118 (2012).
- Fedirko, N., Svichar, N., Chesler, M. Fabrication and use of high-speed, concentric H+- and Ca2+-selective microelectrodes suitable for in vitro extracellular recording. J Neurophys. 96 (2), 919-924 (2006).
- Nicholson, C. Diffusion from an injected volume of a substance in brain tissue with arbitrary volume fraction and tortuosity. Brain Res. 333 (2), 325-329 (1985).
- Nicholson, C., Tao, L. Hindered diffusion of high molecular weight compounds in brain extracellular microenvironment measured with integrative optical imaging. Biophys J. 65 (6), 2277-2290 (1993).
- Thorne, R. G., Nicholson, C. In vivo diffusion analysis with quantum dots and dextrans predicts the width of brain extracellular space. Proc Natl Acad Sci U S A. 103 (14), 5567-5572 (2006).
- Wolak, D. J., Thorne, R. G. Diffusion of macromolecules in the brain: implications for drug delivery. Mol Pharm. 10 (5), 1492-1504 (2013).

