Purification of Low-abundant Cells in the Drosophila Visual System
Summary
Here, we present a cell dissociation protocol for efficiently isolating cells present at low abundance within the Drosophila visual system through fluorescence activated cell sorting (FACS).
Abstract
Recent improvements in the sensitivity of next generation sequencing have facilitated the application of transcriptomic and genomic analyses to small numbers of cells. Utilizing this technology to study development in the Drosophila visual system, which boasts a wealth of cell type-specific genetic tools, provides a powerful approach for addressing the molecular basis of development with precise cellular resolution. For such an approach to be feasible, it is crucial to have the capacity to reliably and efficiently purify cells present at low abundance within the brain. Here, we present a method that allows efficient purification of single cell clones in genetic mosaic experiments. With this protocol, we consistently achieve a high cellular yield after purification using fluorescence activated cell sorting (FACS) (~25% of all labeled cells), and successfully performed transcriptomics analyses on single cell clones generated through mosaic analysis with a repressible cell marker (MARCM). This protocol is ideal for applying transcriptomic and genomic analyses to specific cell types in the visual system, across different stages of development and in the context of different genetic manipulations.
Introduction
The Drosophila visual system is an outstanding model for studying the genetic basis of development and behavior. It comprises a stereotyped cellular architecture1 and an advanced genetic toolkit for manipulating specific cell types2,3. A major strength of this system is the ability to autonomously interrogate gene function in cell types of interest with single cell resolution, using genetic mosaic methods4,5. We sought to combine these genetic tools with recent advances in next generation sequencing to perform cell type-specific transcriptomic and genomic analyses in single cell clones in genetic mosaic experiments.
To accomplish this, it is essential to develop a robust and efficient method to selectively isolate low abundant cell populations in the brain. Previously, we developed a protocol for isolating specific cell types in the visual system during pupal development through FACS, and determining their transcriptomes using RNA-seq6. In these experiments, the vast majority of cells of a particular type (e.g. R7 photoreceptors) were fluorescently labeled. Using this method, in genetic mosaic experiments, wherein only a subset of cells of the same type are labeled, we failed to isolate enough cells to obtain quality sequencing data. To address this, we sought to increase the cellular yield by improving the cell dissociation protocol.
Our approach was to decrease the total length of the protocol to maximize the health of dissociated cells, improve cellular health by altering dissection and dissociation buffers, and reduce the amount of mechanical disruption to the dissected tissue. We tested the improved protocol7 using mosaic analysis with a repressible cell marker5 (MARCM), which allows generation of single fluorescently labeled clones of particular cell types, that are wild type or mutant for a gene of interest in an otherwise heterozygous fly. Where, under identical conditions, our earlier protocol failed to generate enough material for RNA-seq, the improved protocol was successful. We reproducibly achieve a high cellular yield (~25% of labeled cells) and obtain high quality RNA-seq data from as little as 1,000 cells7.
A number of protocols have been previously described to isolate particular cell types in Drosophila6,8,9,10,11,12,13,14,15,16. These protocols are mostly intended for isolating cells that are abundant within the brain. Our protocol is optimized for isolating low abundant cell populations (fewer than 100 cells per brain) in the visual system using FACS for subsequent transcriptomic and genomic analyses. With this protocol, we aim to provide a way to reproducibly isolate low abundant cells from the fly visual system by FACS and obtaining high quality transcriptome data by RNA-seq.
Protocol
1. Planning Before the Experiment
- Before starting the experiment, perform a small-scale pilot experiment to confirm if the genetics work as expected to specifically label the desired cells.
- As a first pass, use immunohistochemistry and light microscopy to determine if unwanted cells are labeled. Ideally, genetic markers will be specific within the retina and optic lobe (Figure 3). Only optic lobes are used for cell dissociation and so labeling in the central brain is not an issue. At the same time, determine how many cells of interest are labeled per brain.
- If the genetic label(s) have the desired specificity, perform a pilot FACS experiment (Figure 4) to determine if a pure population of cells can be reproducibly isolated, and also to assess how many cells of interest can be isolated from each brain. If needed, assess the purity of isolated cells through quantitative PCR to determine the enrichment or de-enrichment of specific genes.
- Determine the number of crosses needed to obtain the desired amount of material for the experiment.
NOTE: Based on experimental experience, ~17–25% of fluorescently labeled cells are consistently isolated with this protocol, and high-quality RNA-sequencing data are obtained from as few as 1,000 FACS purified cells (fewer cells may be sufficient).- To obtain 1,000 cells for RNA-seq analyses, ensure that there are at least 40 cells of interest fluorescently labeled per brain (20 cells per optic lobe). Around ~10 cells of interest should be purified per brain, so dissect 100 brains.
- To obtain ~100 flies at the appropriate stage of development to dissect in each experiment, use ~100 crosses if selecting against balancers (this will vary depending on the genotypes of the parents). If flies are homozygous for the alleles/transgenes of interest fewer crosses can be set.
- Limit the dissection window to 1 h to maximize cellular health and minimize non-specific changes to gene expression and genome organization that can occur after brains are dissected, but this can be adjusted. Determine the number of dissectors required to dissect 100 brains in 1 h, which depends on the skill of the dissectors.
2. Fly Work
NOTE: Flies are raised at 25 °C with ~50% humidity unless otherwise noted. Below the genotype of females is indicated as Genotype F, and that of males as Genotype M.
- Expanding stocks
- Obtain ~100 vials of young flies (no more than a week old) with Genotype F and ~30 vials of Genotype M.
- On Thursday of Week 1, flip 100 vials of Genotype F onto fresh food. Starting on Friday, flip the vials each day through Tuesday of Week 2. These 6 flips will be used to collect virgins. Also on Wednesday of Week 2, flip flies of Genotype M onto fresh food and clear out the parents on Friday of Week 2. These vials will be used to collect males for the crosses.
- Collecting virgins
- From Monday of Week 3, collect virgins from the Genotype F vials. Collect 2-3 times a day and store the virgins at 18 °C with ~50% humidity. Repeat this each day through Tuesday of Week 4. It is highly recommended to collect as many virgins as possible, since a lot of them die before the crosses are set. Typically, ~2,500 virgins should be collected to set 100 crosses.
- Setting crosses
- On Friday of Week 4, set the desired numbers of crosses (typically 100–200 crosses per experiment) with 15 virgins of Genotype F and 7 males of Genotype M for each cross. It is very important to use young and healthy males with intact wings.
- Flip the crosses every day from Sunday through Saturday of Week 5. Supplement the crosses with fresh virgins or males if dead flies are found. It is essential to prevent the food from drying out, so water the vials as needed. Dry food will significantly decrease the yield.
- Negative control for FACS sorting
- Include a negative control without the desired cells being labeled for the FACS sorting. Ideally, the negative control should have the reporter (e.g. UAS-GFP) but not the driver (e.g. GAL4), so that basal expression of the reporter (UAS-constructs can be "leaky") can be determined to set appropriate gating for the FACS sorting. Flip the flies for the negative control at the same time with the experimental flies, and stage them in the same way.
- Staging pupae
- To collect cells at a specific stage of pupal development (e.g. 40 h after puparium formation [h APF]), stage the pupae in a 1 h time window to match with the 1 h period allotted for dissections (discussed above). FACS should be performed directly after cell dissociation, which occurs after dissections and takes ~40 min. Thus, the exact time of staging and dissections is determined by FACS availability.
- To obtain pupae at 40 h APF, stage on Monday of Week 6 at a specific time point (e.g., 5 pm) to dissect pupae 40 h later (e.g., 9 am on Wednesday). Use a wet paintbrush to gently pick white pre-pupae and place them on the wall in pre-warmed vials. Only pick the pupae with desired genotype.
- Biological replicates
- Do at least 3 biological replicates for each experiment. As a simple way, repeat the experiment three times in the same week (Week 6) using the same crosses. For example, stage pupae for the second and third replicates on Tuesday and Wednesday, respectively. Dissect these pupae on Thursday and Friday, respectively.
3. Sample Preparation
NOTE: All the reagents used in this protocol are listed in the Table of Materials.
- Prepare solutions for dissections and tissue dissociation
- Prepare the proteolytic enzyme blend stock solution (see Table of Materials) by reconstituting the lyophilized enzyme with ddH2O to a concentration of 26 Wünsch units (Wu)/mL. For a vial containing 260 Wu (large vial), add 10 mL of ddH2O to the vial, and make single-use aliquots (4.2 µL are required per dissociation). Store stock solution at -20 °C. Each experiment requires dissociation of two samples (negative control and experimental tissue).
- Prepare 10x Rinaldini's Solution (RS) and Complete Schneider's Media (CSM) the day before the dissection (Monday of Week 6). Prepare 5 mL of CSM for each dissector, and about 5 mL of 1x RS and 5 mL of CSM for dissociation of 2 samples (negative control and experimental). Store the 10x RS at 4 °C for up to a month, and CSM at 4 °C for up to a week.
- To prepare 100 mL of 10x RS, dissolve 8 g of NaCl, 200 mg of KCl, 50 mg of NaH2PO4, 1 g of NaHCO3 and 1 g of glucose in 100 mL of ddH2O in a 250 mL beaker. Filter sterilize with a 0.22 mm filter.
- Add 20 mg of L-Glutathione to 1 mL of ddH2O in a microtube.
- To prepare 50 mL of CSM, add 5 mL of heat inactivated bovine serum, 0.1 mL of 10 mg/mL insulin solution, 5 mL of 200 mM L-Glutamine solution, 12.5 μL of 20 mg/mL L-Glutathione, and 1 mL of Penicillin-Streptomycin solution (5,000 units of penicillin and 5 mg of streptomycin/mL) to 38.9 mL of Schneider's media. Filter sterilize with a 0.22 mm filter.
- Dilute 10x RS with ddH2O to make 1x RS working solution. Prepare 500 μL of 1x RS in non-adhesive microtubes for each sample.
- Turn on a water bath and set the temperature at 37 °C. Make sure the temperature is accurate by using a thermometer. The water bath must be at the correct temperature prior to papain activation. It is recommended to set the temperature the evening before a morning dissection.
- Prepare papain and proteolytic enzyme blend
NOTE: Make papain solution fresh before every dissociation.- On Tuesday of Week 6 around 45 min before starting the dissections, retrieve 1 vial of papain powder from 4 °C and incubate at room temperature. Also set aside CSM in an Eppendorf tube at room temperature to later add to the papain powder.
- min prior to the start of dissections use a syringe to add the room temperature CSM to the vial of papain (directly through the cap) so that the concentration is 100 units/mL (the amount of powder in each vial is slightly different; for a vial containing 123 units add 1.23 mL of CSM). To mix, first invert the vial to capture any powder stuck on the inside of the cap, and then pipette up and down.
- Add the vial to a beaker containing 37 °C water in the water bath. Make sure the vial of papain solution is not floating. Activate the papain for 30 min at 37 °C.
- After adding the papain to a beaker in the water bath, retrieve an aliquot of proteolytic enzyme blend (26 Wu/mL) from -20 °C and incubate it at room temperature. After 30 min of activation, incubate the papain at room temperature.
- Dissections
- On Tuesday of Week 6, dissect the staged pupal brains in cold CSM with clean forceps, and cut off the optic lobes by pinching at the junction of the optic lobe and the central brain.
- Add the lobes to the bottom of a microtube containing 500 μL of 1x RS (one microtube for the experimental tissue and one for the negative control tissue). Always keep the tubes on ice.
- Dissect as many lobes as possible in 1 h. For the negative control tissue, 10 lobes are enough. Pool dissected optic lobes into a single microtube to eliminate tissue loss during transfer between microtubes.
- Cell dissociation
- Carefully remove the 1x RS from the microtube with dissected optic lobes using a P200 pipette, leaving enough to cover the lobes. Carefully add 500 μL of 1x RS to the side of the tube, so as to disturb the brains as little as possible. Always keep the tube on ice.
- Let the lobes settle to the bottom. Just once, take up 200 μL and pipette out to mix (lobes should move around). Let the lobes settle again.
- For two samples (experimental and negative control), aliquot 600 μL of activated room temperature papain into a microtube. Add 4.2 μL of room temperature proteolytic enzyme blend into 600 μL papain solution to obtain a final concentration of 0.18 Wu/mL. Mix by pipetting up and down. This is the dissociation solution.
- Carefully remove the 1x RS from each sample and add 300 μL of dissociation solution to the lobes. Incubate the samples at 25 °C, 1,000 rpm for 15 min in a microtube thermomixer.
- At the 5 and 10 min time points, stop the thermomixer and let the lobes sink to the bottom of the microtube. Take up 200 μL of solution and pipet out to mix.
- After 15 min, let the lobes settle to the bottom and carefully remove the dissociation solution from the lobes (try not to disturb the lobes).
- Wash twice with 1x RS. To do this, carefully add 500 μL 1x RS (try not to disturb the lobes). To mix, gently pipet up 200 μL, and then pipet out into the microtube. Let the lobes sink to the bottom and repeat.
- Wash each sample twice with 500 μL of CSM (same as previous step).
- Carefully remove the CSM and add another 200 μL of CSM to the tube. Using a P200 pipette, disrupt the tissue by pipetting up and down without foaming, until the solution is homogenous (i.e., can no longer see any remnants of tissue).
- Filter the cell suspension through a 30 μm mesh cap into a 5 mL FACS tube on ice (use a P200 and make sure the entire suspension goes through). Add another 500 μL of CSM to rinse the dissociation microtube, and filter the solution into the FACS tube.
- Carefully take off the cap of the FACS tube, collect the solution underneath the mesh filter with a P200 and add it to the rest of the suspension in the FACS tube. The samples are ready for FACS.
- Be sure to have microtubes for collecting FACS purified cells ready in advance. For RNA-seq, sort cells from each sample directly into microtubes containing 300 μL of RLT buffer (lysis buffer) from the RNA purification kit (add 1:100 2–Mercaptoethanol before use).
- FACS sorting
NOTE: After dissociation, immediately sort the cells by FACS (1 h max). Make sure to book FACS sessions accordingly when planning for the experiment. A cell sorter with 100 μm nozzle size is typically used.- Compare the cell distribution in the negative control and the experimental sample to set up the gating for the sorting. Ideally, establish the gating in the pilot FACS experiment, and the negative control sample will provide confirmation or fine tuning of the pre-established gating. The cells of interest should be well separated from the background cells that are present in both the negative control and the experimental sample (Figure 4) (test this in the pilot experiments).
- Collect the sorted cells in the prepared collection tubes.
- RNA purification
- After sorting the cells directly into 300 μL of RLT buffer, pipet up and down to lyse the cells.
- Purify RNA following the standard protocol of Purification of Total RNA from Animal and Human Cells using the purification kit (see Table of Materials) with on-column DNA digestion7. Elute the RNA in 20 μL of RNase-free water.
- Freeze the RNA samples with liquid nitrogen and store at -80 °C.
- For each experiment perform at least 3 replicates for control and experimental conditions. Prepare the cDNA libraries for all samples at the same time and run all the samples on the same flow cell to control for technical variability. Once all the RNA samples are prepared, use a speed vac to reduce the volume to 2–5 μL (each sample).
4. Preparing cDNA Libraries and Sequencing by Smart-seq2
NOTE: To minimize technical variability, make all the cDNA libraries at the same time, and sequence them on the same flow cell.
- Use an input of 1.5 μL of concentrated RNA to make cDNA libraries by following the standard protocol for Smart-seq217 except for the following modifications: use a different high-fidelity PCR master mix (see Table of Materials) for PCR amplification; use an on-column PCR purification kit (see Table of Materials) instead of magnetic beads to purify PCR products.
- The Smart-seq2 protocol includes a PCR pre-amplification step after reverse transcription as well as a final enrichment PCR step after tagmentation. The number of cycles in the PCR amplification depends on the abundance of the input material. With 1,000 cells as input, typically use 15 cycles in the PCR pre-amplification step and 12 cycles for the final enrichment PCR step.
- Pool the libraries and sequence them on a single flow cell (e.g., the NextSeq platform).
Representative Results
General scheme
A general scheme of the protocol is shown in Figure 1. The protocol is divided into three major parts: fly work, sample preparation, sequencing and data analysis. The "Planning before the experiment" session of the protocol is not included in the general scheme for simplicity.
Timeline
A calendar of the major parts of the protocol (Fly work and Sample preparation) is shown in Figure 2.
Verifying cell-specific labeling by confocal microscopy
In a representative experiment, L3 lamina monopolar neurons (L3) were fluorescently labeled. L3 neuron is one of the five homologous lamina neurons (L1-L5) that receive input from the broadly tuned photoreceptors R1-R6 and relay the information to the high center by synapsing to target neurons in the medulla. In a MARCM experiment, single cell L3 clones are generated by mitotic recombination and fluorescently labeled by two markers, myr-tdTOM (membrane) and H2A-GFP (nuclear). The genotypes of flies used in the crosses are shown in Table 1. To verify cell-specific labelling, fly brains of the desired genotype were dissected at the developmental stage of interest (40 h APF). Immunostaining was performed as previously described7 using antibodies specific against dsRed and GFP (Figure 3, magenta and green), as well as the 24B10 antibody as a reference for the lamina and medulla neuropils (Figure 3, blue). Confocal microscopy confirmed that L3 is the only cell type labelled by both dsRed and GFP in the optic lobe.
Isolation of low-abundance cells by FACS
A representative FACS sorting result is shown in Figure 4. 100,000 events were recorded. 29.9% of all the events are potential singlets, and the rest could be debris or aggregates. Doublets and cells with different sizes were then gated out based on granularity and size. From the remaining single cells (FSC singlets, 27.3% of all events), L3 neurons appeared as a tight cluster in P1 (Figure 4, Specimen, magenta), which was well separated from the background cells (Figure 4, Specimen, blue). The size of the cells in P1 was similar consistent with a homogenous cellular population. Notably, a few cells in P2 (Figure 4, Specimen, green) with intermediate fluorescence intensity of dsRed and GFP were distributed between the tight cluster of cells in P1 and the background. The identity of these cells was not clear. Since P2 cells had a different size than the P1 cells, these cells were likely to be non-specific. To avoid potential contamination from the non-specific cells, only P1 cells were collected for the experiment. From 100,000 events, 45 P1 cells are obtained.
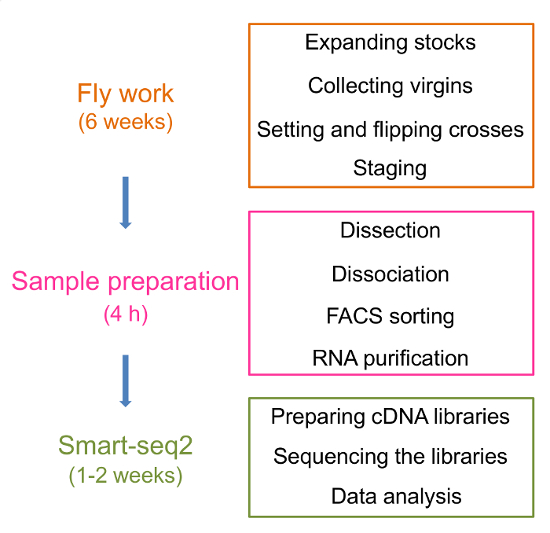
Figure 1: A general scheme of the protocol. The protocol consists of three parts: fly work, sample preparation, sequencing and data analysis. The approximate processing time of each part is indicated. Major steps of each part are also shown in the corresponding boxed regions. Please click here to view a larger version of this figure.
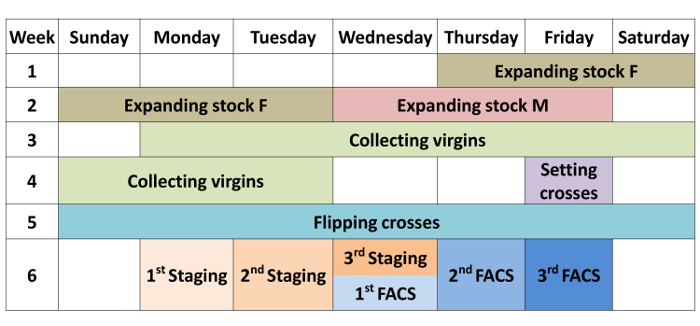
Figure 2: Timeline for fly work and Sample preparation. The protocol takes about 6 weeks. Timing for the major steps is shown in the calendar. Dissection, dissociation and RNA purification are done in the same day as the FACS sorting, and are not shown in the calendar for simplicity. Three biological replicates are done sequentially in the same week with the same crosses. Please click here to view a larger version of this figure.
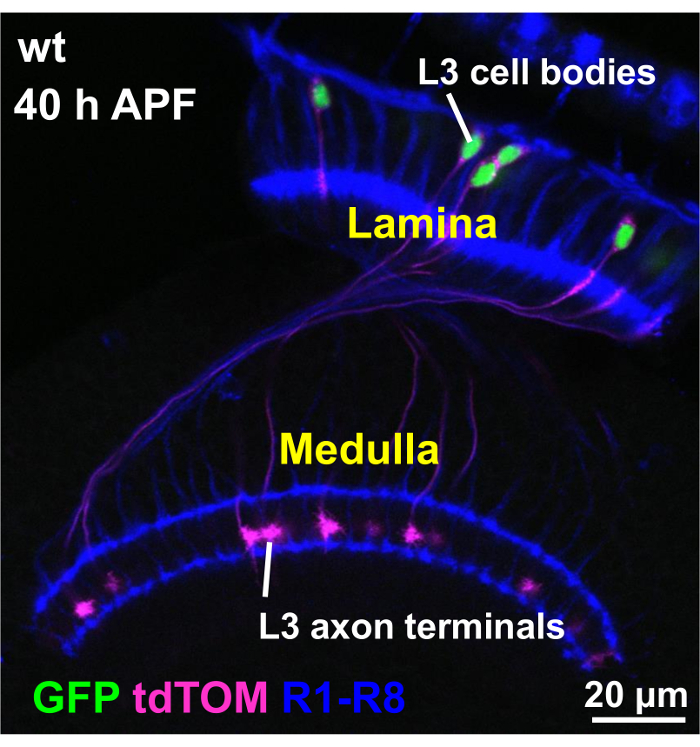
Figure 3: A representative confocal microscopy image showing the selective labeling of desired cells by fluorescent markers. In MARCM experiments, single L3 lamina monopolar neurons were made to express myr-tdTOM and H2A-GFP using an L3-specific GAL4 driver (9-9-GAL4)18. Fly brains of the desired genotype were dissected at 40hr APF and stained with anti-dsRed, anti-GFP and 24B10 antibodies (as a reference for the lamina and medulla neuropils). Fluorescent labeling was assessed by confocal microscopy. L3 neurons are born in the lamina and project axons that terminate within the medulla neuropil. In each brain, a subset of L3 neurons expressed both fluorescent reporters, and these were the only cells in the optic lobe expressing the markers. Please click here to view a larger version of this figure.
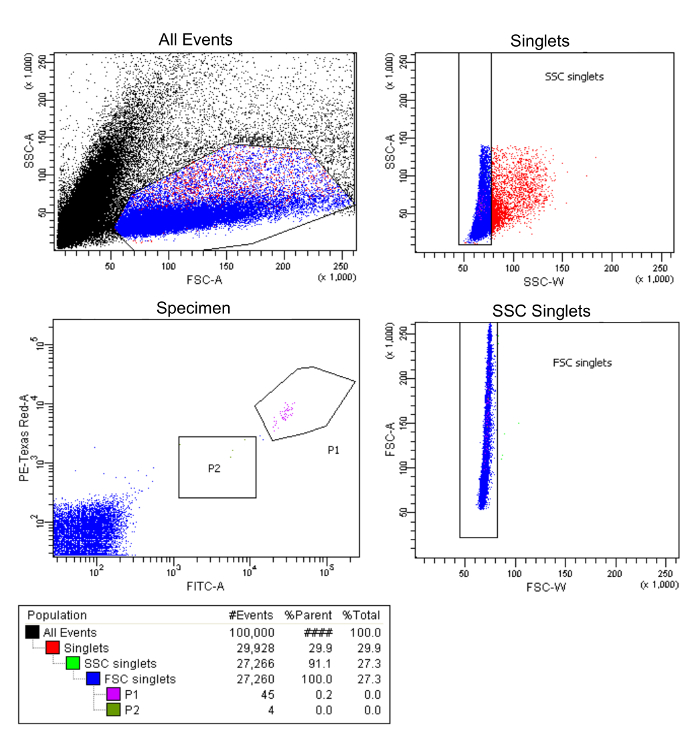
Figure 4: Purifying single L3 lamina neuron MARCM clones via FACS: a representative FACS plot. Gates (e.g. P1) were created based on cell granularity, size, and fluorescence intensity to isolate homogeneous single cells. L3 neurons expressing similarly high levels of myr-tdTOM and H2A-GFP were collected from in P1 (magenta). A few cells with intermediate fluorescence intensity are observed in P2 (green). These are different in size than L3 neurons in P1, and could represent non-specific cells. These were not collected. Please click here to view a larger version of this figure.
| Genotype | Source |
| w; TubP-GAL80, FRT40, 27G05-FLP::PEST/CyO, Kr-GAL4, UAS-GFP; 9-9-GAL4, UAS-myr::tdTOM/TM6B | Peng et al., 2018 |
| w; FRT40/CyO, Kr-GAL4, UAS-GFP; UAS-H2A-GFP/TM6B | Peng et al., 2018 |
Table 1: Genotypes of flies used in the crosses.
Discussion
This protocol is simple and not technically difficult to execute, but there are several key steps that if overlooked will cause a considerable reduction in cellular yield. (Step 2.3.2.) It is crucial that crosses are healthy, and that the food does not dry out. Regular watering of crosses is essential to maximize the number of flies available for dissection that are of the right genotype and at the correct stage of development. How often crosses need to be watered will vary depending on the food used and the housing conditions of the flies. To ensure crosses don't dry out, examine the vials twice a day and add water accordingly. (Step 3.3.) It is also important to pool dissected brains into a single microtube that will be used for the dissociation, as opposed to having each dissector use their own microtube and then subsequently pooling the samples. This will eliminate any brains lost from transferring between microtubes, which can be considerable. (Step 3.4.9.) When pipetting up and down to manually disrupt tissue, it is vital to do so until no remnants of the tissue are visible by eye in the suspension. Incomplete tissue disruption will reduce the number of single cells that can be sorted via FACS, and thus decrease cellular yield.
The major limitation of cellular yield is starting material. In this regard, the dissections are rate limiting for several reasons: (1) the dissociation protocol requires optic lobes to be dissected out of the head and separated from the central brain, (2) to obtain tissue at precise time-points during development the time window for dissections must be limited, (3) The longer the time-period between dissections and FACS, the greater the chance of suboptimal cell health and non-specific effects on gene expression and genomic organization (i.e., the shorter the dissection period the better). The time spent dissecting should be modified to obtain the desired amount of material in the minimal amount of time. Most of the troubleshooting is performed in the pilot experiments. Here it is crucial to: optimize genetic labeling for brightness and specificity (Figure 3), assess whether a reproducible population of cells of interest are clearly segregated from background cells in FACS plots (Figure 4), determine the number of brains that need to be dissected to obtain a sufficient amount of material for subsequent applications (see Step 1. Fly work), and determine the minimal amount of time needed to dissect the appropriate numbers of brains.
The major advance of this protocol is that it allows efficient isolation of low abundant cells for transcriptomic and genomic applications, improving sensitivity considerably over our previous method6. Based on our estimations, fluorescently labeled populations comprising fewer than 100 cells per optic lobe can be efficiently isolated for RNA-seq analyses. This allows transcriptomic and genomic analyses to be applied in genetic mosaic experiments, wherein subsets of cells of particular types are genetically manipulated in an otherwise normal background. This represents a critical advance, as such experiments are essential for determining cell autonomous and non-autonomous gene functions in complex tissues.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was funded by the NINDS of the National Institutes of Health under award number K01NS094545, andgrants from the Lefler Center for the Study of Neurodegenerative Disorders. We acknowledge Liming Tan and Jason McEwan for valuable conversations.
Materials
| Liberase TM | Roche | 5401127001 | Proteolytic enzyme blend |
| NaCl | Sigma-Aldrich | S3014 | |
| KCl | Sigma-Aldrich | P9541 | |
| NaH2PO4 | Sigma-Aldrich | S9638 | |
| NaHCO3 | Sigma-Aldrich | S5761 | |
| Glucose | Sigma-Aldrich | G0350500 | |
| L-Glutathione | Sigma-Aldrich | G6013 | |
| Heat Inactivated Bovine Serum | Sigma-Aldrich | F4135 | |
| Insulin Solution | Sigma-Aldrich | I0516 | |
| L-Glutamine | Sigma-Aldrich | G7513 | |
| Penicillin-Streptomycin Solution | Sigma-Aldrich | P4458 | |
| Schneider's Culture Medium | Gibco | 21720024 | |
| Papain | Worthington | LK003178 | |
| 2-Mercaptoethanol | Sigma-Aldrich | M6250-100ML | |
| RNeasy Micro Kit | Qiagen | 74004 | RNA purification kit |
| RNase-free DNase | Qiagen | 79254 | |
| SuperScript II Reverse Transcriptase | Life Technologies | 18064-014 | |
| dNTP Mix | Life Technologies | R0191 | |
| MgCl2 Solution | Sigma-Aldrich | M1028-10X1ML | |
| Betaine Solution | Sigma-Aldrich | B0300-1VL | |
| RNaseOUT | Life Technologies | 10777-019 | |
| Q5 High-Fidelity 2x Master Mix | New England Biolabs | M0492S | |
| MinElute PCR Purification Kit | Qiagen | 28004 | |
| Nextera XT DNA Library Prepration Kit | Illumina | FC-131-1024 | |
| Nextera XT Index Kit | Illumina | FC-131-1001 | |
| Test Tube with Cell Strainer Snap Cap | Falcon | 352235 | |
| Bottle-Top Vacuum Filter Systems | Corning | CLS431153 | |
| ThermoMixer F1.5 | Eppendorf | 5384000020 | |
| FACSAria Flow Cytometer | BD Biosciences | 656700 | |
| HiSeq 2500 Sequencing System | Illumina | SY–401–2501 |
References
- Fischbach, K. -. F., Dittrich, A. P. M. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell and Tissue Research. 258, 441-475 (1989).
- Pfeiffer, B. D., et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 105 (28), 9715-9720 (2008).
- Jenett, A., et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2 (4), 991-1001 (2012).
- Xu, T., Rubin, G. M. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117 (4), 1223-1237 (1993).
- Lee, T., Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 22 (3), 451-461 (1999).
- Tan, L., et al. Ig Superfamily ligand and receptor pairs expressed in synaptic partners in drosophila. Cell. 163 (7), 1756-1769 (2015).
- Peng, J., et al. Drosophila Fezf coordinates laminar-specific connectivity through cell-intrinsic and cell-extrinsic mechanisms. Elife. 7, (2018).
- Krasnow, M. A., Cumberledge, S., Manning, G., Herzenberg, L. A., Nolan, G. P. Whole animal cell sorting of Drosophila embryos. Science. 251 (4989), 81-85 (1991).
- Cumberledge, S., Krasnow, M. A. Preparation and analysis of pure cell populations from Drosophila. Methods in Cell Biology. 44, 143-159 (1994).
- Bryant, Z., et al. Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis. Proceedings of the National Academy of Sciences of the United States of America. 96 (10), 5559-5564 (1999).
- Neufeld, T. P., de la Cruz, A. F., Johnston, L. A., Edgar, B. A. Coordination of growth and cell division in the Drosophila wing. Cell. 93 (7), 1183-1193 (1998).
- Calvi, B. R., Lilly, M. A. Fluorescent BrdU labeling and nuclear flow sorting of the Drosophila ovary. Methods in Molecular Biology. , 203-213 (2004).
- Tirouvanziam, R., Davidson, C. J., Lipsick, J. S., Herzenberg, L. A. Fluorescence-activated cell sorting (FACS) of Drosophila hemocytes reveals important functional similarities to mammalian leukocytes. Proceedings of the National Academy of Sciences of the United States of America. 101 (9), 2912-2917 (2004).
- Bryantsev, A. L., Cripps, R. M. Purification of cardiac cells from Drosophila embryos. Methods. 56 (1), 44-49 (2012).
- Harzer, H., Berger, C., Conder, R., Schmauss, G., Knoblich, J. A. FACS purification of Drosophila larval neuroblasts for next-generation sequencing. Nature Protocols. 8 (6), 1088-1099 (2013).
- Khan, S. J., Abidi, S. N., Tian, Y., Skinner, A., Smith-Bolton, R. K. A rapid, gentle and scalable method for dissociation and fluorescent sorting of imaginal disc cells for mRNA sequencing. Fly (Austin). 10 (2), 73-80 (2016).
- Picelli, S., et al. Full-length RNA-seq from single cells using Smart-seq2. Nature Protocols. 9 (1), 171-181 (2014).
- Nern, A., Zhu, Y., Zipursky, S. L. Local N-cadherin interactions mediate distinct steps in the targeting of lamina neurons. Neuron. 58 (1), 34-41 (2008).

