Use of Bisection to Reduce Mitochondrial DNA in the Bovine Oocyte
Summary
Here, we present a protocol to significantly reduce the mitochondrial DNA copy numbers in a bovine oocyte (P < 0.0001). This method utilizes centrifugation and bisection to substantially reduce oocyte mitochondria and may allow for an increased chance of development in the reconstructed interspecies somatic cell nuclear transfer embryos.
Abstract
Interspecies somatic cell nuclear transfer (iSCNT) may be used to rescue endangered species, but two distinct populations of mitochondrial DNA (mtDNA) exist within the reconstructed embryo: one within the recipient ooplasm and one within the donor somatic cell. This mitochondrial heteroplasmy can lead to developmental issues in the embryo and the fetus. Handmade cloning protocols include oocyte bisection, which can be used to decrease the mtDNA copy number, reducing the degree of mitochondrial heteroplasmy in a reconstructed embryo. Centrifugation of denuded, mature bovine oocytes produced a visible mitochondria-dense fraction at one pole of the oocyte. Oocytes' zonae pellucidae were removed by exposure to a pronase solution. Bisection was performed using a microblade to remove the visible mitochondria fraction. qPCR was used to quantify the mtDNA present in DNA samples extracted from whole oocytes and bisected ooplasts, providing a comparison of mtDNA copy numbers before and after bisection. Copy numbers were calculated using cycle threshold values, a standard curve's regression line formula, and a ratio that included the respective sizes of mtDNA PCR products and genomic PCR products. One bovine oocyte had an average mtDNA copy number (± standard deviation) of 137,904 ± 94,768 (n = 38). One mitochondria-depleted ooplast had an average mtDNA copy number of 8,442 ± 13,806 (n = 33). Average mtDNA copies present in a mitochondria-rich ooplast were 79,390 ± 58,526 mtDNA copies (n = 28). The differences between these calculated averages indicate that the centrifugation and subsequent bisection can significantly decrease the mtDNA copy numbers present in the mitochondria-depleted ooplast when compared to the original oocyte (P < 0.0001, determined by one-way ANOVA). The reduction in mtDNA should decrease the degree of mitochondrial heteroplasmy in a reconstructed embryo, possibly fostering standard embryonic and fetal development. Supplementation with mitochondrial extract from the somatic donor cell may also be essential to achieve successful embryonic development.
Introduction
Somatic cell nuclear transfer (SCNT) includes the fusion of an enucleated oocyte from one animal and a somatic cell from an animal of the same species. In most cases, the oocyte and somatic cell originate from the same species, and live birth rates are below 6%1. Some research involves the use of interspecies SCNT (iSCNT), which includes the fusion of a somatic cell and oocyte that originate from two different species. In these studies, live birth rates are even lower than in SCNT-typically less than 1%1. However, iSCNT has the capacity to be used as a method of rescuing endangered species, since somatic cells from these animals are more accessible than their germ cells1. Recipient oocytes used in iSCNT are often domestic or common laboratory species, such as cows, pigs, and mice. Some attempts made thus far have successfully produced live young, though the offspring produced have been intrageneric animals (the recipient oocyte species and donor cell species were members of the same genus)2,3,4. Intergeneric models (which utilize an oocyte and somatic cell from animals in different genera) have not yet produced live animals, and the majority of reconstructed embryos arrest at the 8-16 cell stage of in vitro development5,6,7,8. One possible explanation of this embryonic developmental arrest is the occurrence of mitochondrial heteroplasmy in the embryos-the presence of more than one mitochondria DNA (mtDNA) type in a single cell. Heteroplasmy can lead to issues such as developmental inefficiency or failure in the embryo or in the live animal1. Pathogenesis can also occur later in the animal's lifetime9. Though this issue is also present in SCNT offspring, the interspecific component within iSCNT embryos exacerbates the issue.
When the embryonic mtDNA comes from two different species, the recipient oocyte mitochondria, which represent the majority, do not work efficiently or effectively with the donor cell's nucleus1,10. Larger taxonomic gaps between the two species used in iSCNT likely intensifies this problem; intrageneric live offspring produced (Bos gaurus and Bos indicus offspring using Bos taurus oocytes), as well as offspring produced via traditional SCNT (e.g. Ovis aries offspring using Ovis aries oocytes) were shown to be chimeras (mtDNA from two individuals was present in these animals11,12,13). Yet, they developed much further than the intergeneric SCNT embryos14,15. The exchange of information between the oocyte mitochondria and the donor cell's nucleus could be more successful in the intrageneric embryo than in the intergeneric embryo16.
The amount of mtDNA in a mature bovine oocyte is approximately 100 times greater than the amount found in one somatic cell12. Reducing this ratio could encourage the somatic cell mitochondria to proliferate within the reconstructed embryo, allowing for a greater population of productive mitochondria to be present16. This could in turn provide more energy to meet the requirements of the developing embryo15. Previous attempts made to reduce the mtDNA copy number of the oocyte or embryo include chemical application, micromanipulation, and supplementing the oocyte or embryo with additional mitochondria from the donor cell species16,17,18,19,20. However, chemical application (such as 2',3'-dideoxycytidine) is not ideal for embryonic development, and has reduced oocyte mtDNA copy numbers by approximately half18. Prior oocyte mtDNA reduction by micromanipulation have only removed an average of 64% of the oocyte's mtDNA17. Though the supplementation of donor cell mitochondria could be a viable option, its use has not yet produced a live intergeneric animal within iSCNT studies21.
The use of bisection to reduce oocyte mtDNA copy number has not yet been used in published studies. Bisecting oocytes with the intention of fusing the ooplasts with a somatic cell is the premise of handmade cloning (HMC), which typically utilizes bisection as method of removing the polar body and metaphase plate from the metaphase II (MII) oocyte. HMC has successfully produced offspring in several species, including goats, cattle, pigs, sheep, and horses22,23,24,25,26, but does not typically include a centrifugation step prior to bisection. Integrating high-speed centrifugation of the oocyte allows for the isolation of mitochondria (and therefore mtDNA) at one pole of the oocyte, which can then be bisected using a microblade to remove those mitochondria-dense fractions. Two mitochondria-depleted ooplasts can then be fused with a somatic cell, as is the case in HMC, to form a reconstructed embryo which contains considerably less mtDNA from the oocyte species.
The question we attempt to answer with this protocol is how to reduce mtDNA in the bovine oocyte in order to produce a viable reconstructed embryo that contains less heteroplasmic mtDNA. In this protocol, oocytes were centrifuged and bisected. Ooplast and intact oocyte mtDNA copy numbers were calculated to determine the effectiveness of this technique in reducing the bovine oocyte's mtDNA copy number.
Protocol
The following protocol follows the animal care and ethics guidelines provided by Utah State University.
1. Media preparation
- Prior to oocyte handling, prepare the following solutions, as described in Table 1: 400 µL of Hyaluronidase Solution, 500 µL of T2 media, 1,020 µL of T20 media, and 800 µL of T10 media.
- Divide the T10 media into two wells of a four-well plate, 400 µL per well. Label one well with "M" and the second well with "MR." Place in a 5% CO2 incubator until after bisection.
- Prepare 500 µL of Cytochalasin B (CB)/Synthetic Oviductal Fluid with HEPES (HSOF). Aliquot 50 µL of CB/HSOF solution into a separate 1.5 mL centrifuge tube.
- Prepare 40 µL of Pronase Solution. Centrifuge for at least 30 s at 2,680 x g. Mix 20 µL of the supernatant with 20 µL of T2 to a new centrifuge tube; this forms a diluted, 5 mg/mL pronase solution.
- Prepare 3 mL of HSOF. On a search plate, separately deposit four 400 µL drops; place this plate under a stereomicroscope.
- Prepare 500 µL of CB/T20 solution.
2. In vitro maturation (IVM) of bovine oocytes
- IVM: Culture aspirated cumulus-oocyte complexes (COCs) at 38.5 °C in a 5% CO2 incubator for 21 h in a four-well dish of maturation media containing 10% FBS, 0.26 IU/mL FSH, and 100 U/mL penicillin/streptomycin.
- Perform the following steps to denude the oocytes.
- Collect the desired number of the COCs using a 200 µL pipette, and deposit them at the bottom of a 1.5 µL centrifuge tube.
- Add the same volume of 0.6 mg/mL hyaluronidase to the centrifuge tube with the oocytes (i.e., if the volume of oocytes and IVM media is 100 µL, add 100 µL of hyaluronidase).
- Pipette the solution up and down without creating bubbles, until all the cumulus cells have been removed.
- Check the maturation of the oocytes.
- Use the pipette to add 200 µL of HSOF from one of the four HSOF drops on the search plate to the oocyte/hyaluronidase solution.
- Transfer the oocytes from the hyaluronidase/HSOF drop and place them in unused 400 µL of HSOF drop. Wash them with two additional HSOF drops to assist in removing hyaluronidase and cumulus cell remnants.
- Using a mouth pipette (or a 10 µL pipette and tip) and high microscopic magnification, roll and select the oocytes based on polar body presence.
- After sorting the oocytes, collect the desired number of MII oocytes to be bisected, and place them in 400 µL of HSOF drop.
NOTE: If the oocytes have been outside of the incubator for longer than 30 min, oocytes may be placed back into a well of maturation media and placed in a CO2-controlled incubator to rest for at least 30 min. Limit the number of oocytes to be bisected to an amount that is feasible to finish bisecting in approximately 30 min to minimize time outside of incubation. If ooplasts are to be fused with a somatic cell, activated, and cultured as embryos, oocytes should be enucleated at this time.
3. Centrifugation of the oocytes
NOTE: If oocytes were placed in the incubator in maturation media, move them to the HSOF drop from which they were most recently collected.
- Using a mouth pipette, collect the selected mature oocytes from the HSOF drop, and place them into the 1.5 mL tube containing 50 µL of the HSOF/CB solution. Centrifuge the oocytes at 15,000 x g for 12 min.
NOTE: It is not detrimental for bubbles to be present in the centrifuge tube. - While the oocytes are being centrifuged, prepare the bisection plate.
- Make the pattern as shown in Figure 1 on the lid of a 60 mm petri dish (20 µL per drop), and fully cover the drops with mineral oil.
- Using a thin-tipped marker, mark the lines underneath the dish, as shown in Figure 2.
- Draw a box around the pronase drop and a line between the CB/T20 drops and the bottom row of T20 drops.
- Cover the dish with an opaque lid until centrifugation is complete to prevent osmolarity changes of microdrops.
- As soon as centrifugation is complete, use a 200 µL pipette to collect the oocytes within the solution, and move them into an empty portion of a new search plate with four 400 µL HSOF drops.
- Collect and wash the oocytes through the four HSOF drops.
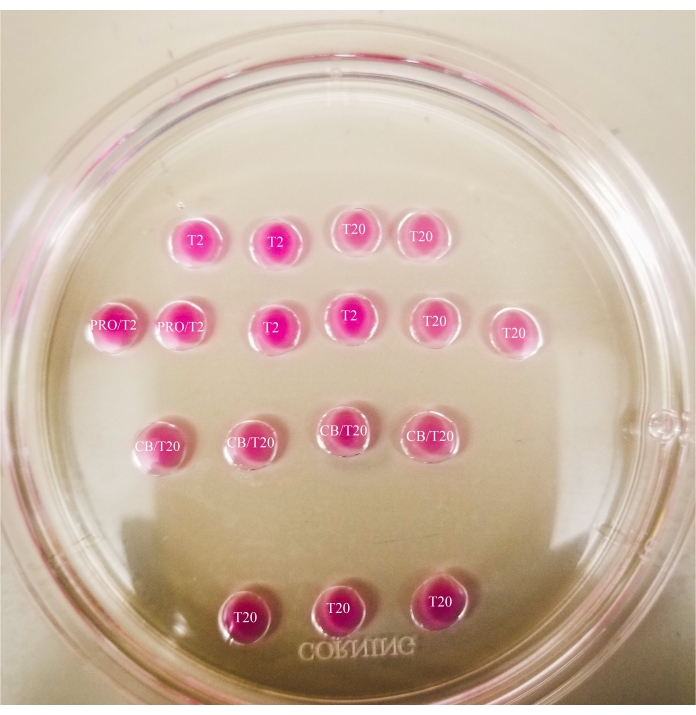
Figure 1: Bisection plate. All drops shown have a 20 µL volume. The plate has a diameter of 60 mm. Drops have been completely covered with mineral oil. Oocytes will first be placed in the uppermost and leftmost T2 drop (indicated here with a star). PRO/T2: 10 µL of pronase and 10 µL of T2, combined prior to creating microdrops. CB/T20: 1 µL of cytochalasin B per 1 mL of T20, combined prior to creating microdrops. Please click here to view a larger version of this figure.
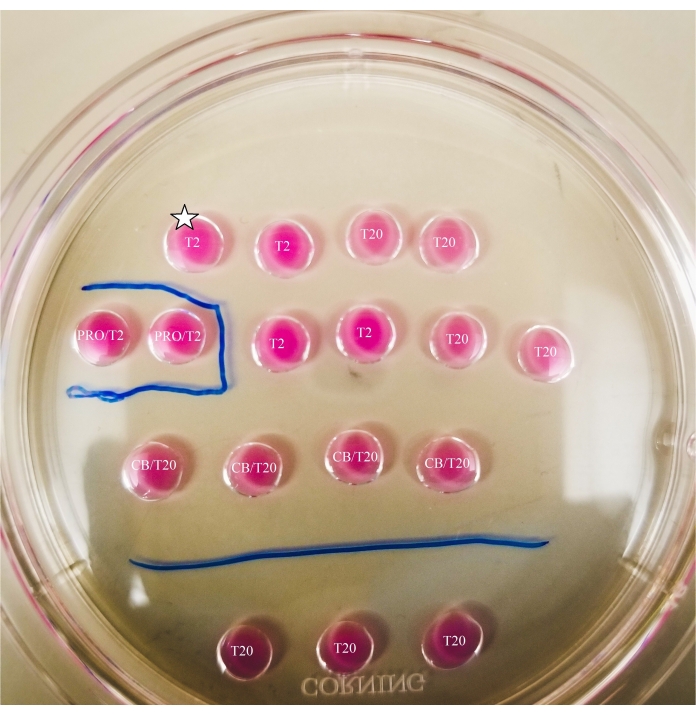
Figure 2: Marked bisection plate. Lines are drawn with a thin-tipped marker on the bottom of the plate, to provide location references for observations and oocyte and ooplast transfers made underneath the microscope. Oocytes will first be placed in the uppermost and leftmost T2 drop (indicated here with a star). Please click here to view a larger version of this figure.
4. Preparation of the oocyte for bisection
NOTE: The following process involves preparation of the oocytes for bisection.
- Turn the heating stage on to 37 °C.
- Use a mouth pipette to move the centrifuged oocytes from the HSOF drop to the top left T2 drop of the bisection plate, then wash the oocytes through the next three drops on the top row (T2, T20, T20).
- Deposit the oocytes in one of the pronase drops, assuring there is little to no contact between them.
NOTE: The removal of the zona pellucida can take varying amounts of time, but typically occurs between 30-120 s with the use of a heating stage. The absence of a heating stage will extend this time. - Observe the oocytes until there is clear deformation of the zonae pellucidae (see Figure 3).
- Once a single oocyte zona pellucida has become deformed, move that oocyte to the neighboring T2 drop. Repeat as additional oocytes become deformed, until all oocytes have been removed from the pronase and placed in the T2 drop.
- Observe the oocytes until only a thin layer of the zone pellucida remains.
- Wash the oocytes through the next three drops within the row (T2, T20, T20), then deposit them in vertical lines within the CB/T20 drops (see Figure 4).
- Begin with fewer oocytes in the vertical lines and increase the number of oocytes per drop as bisection skills progress.
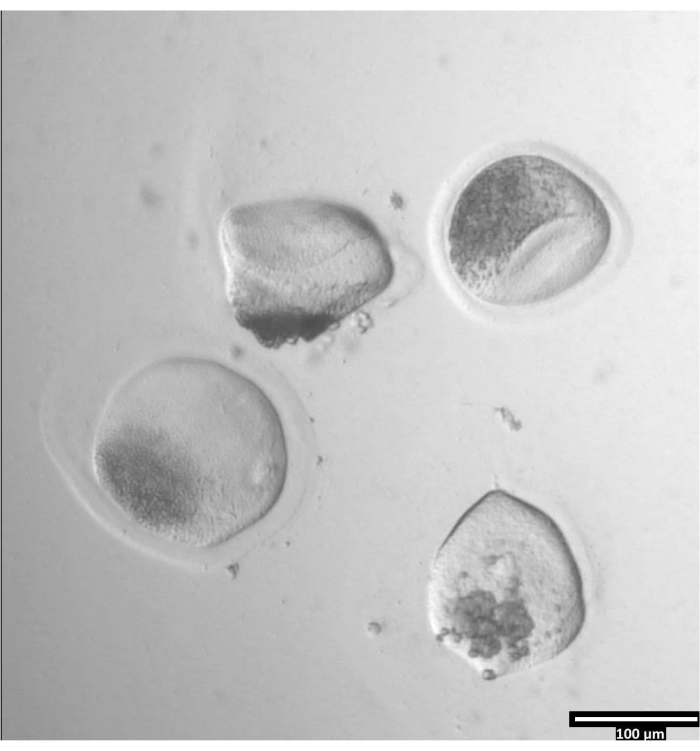
Figure 3: Removal of zona pellucida using pronase. (80x) An oocyte zona pellucida will begin to appear deformed when the pronase has affected the zona pellucida enough for the oocyte to be moved to the adjacent T2 drop. Please click here to view a larger version of this figure.

Figure 4: Oocyte orientation in bisection drops. (80x) Zona-free oocytes are deposited in a near-vertical orientation within each CB/T20 drop prior to bisection. Please click here to view a larger version of this figure.
5. Bisection of the oocytes
- Focus on the first, left-most CB/T20 (bisection) drop that contains oocytes, and use a mouth pipette to rotate the oocytes, so that the mitochondria-dense portion of each oocyte is either facing toward or away from the microscope's arm.
NOTE: The mitochondria-dense portion will not contain lipids, which appear as the darkest portion of the oocyte following centrifugation. - Rest the tip of the microblade to the left of the topmost oocyte, in line with the space directly above the mitochondria-dense portion. Keeping the tip of the blade in the same place, carefully lower the blade to cut all the way through the oocyte.
- Ensure that the mitochondria-dense ooplast and mitochondria-reduced ooplast are similar in size; bisection should cut each oocyte in half to produce two ooplasts.
NOTE: Comparative sizes of ooplasts produced from a single oocyte may vary based on research goals; if mitochondria-reduced ooplasts are to be fused with a somatic cell, their volume should be greater than the mitochondria-dense ooplasts'. - Ensure that the blade is bisecting the oocyte above the mitochondria-dense portion and below the lipid-dense portion, in the clearest segment of the centrifuged oocyte.
- Ensure that the mitochondria-dense ooplast and mitochondria-reduced ooplast are similar in size; bisection should cut each oocyte in half to produce two ooplasts.
- When lifting the blade, ensure that the same line is maintained, and that the tip of the blade remains in same place, then gently lift the tip from the plate.
- Repeat bisection steps for all oocytes within the first bisection drop. If oocytes are present in additional drops, orient and bisect the remaining oocytes.
- Using a mouth pipette, collect mitochondria-reduced ooplasts from the first bisection drop. Place them in the left-hand T20 drop located in the bottom row of the plate. Repeat for all remaining bisection drops.
NOTE: Mitochondria-reduced ooplasts will contain lipids, that are darker in color than the rest of the oocyte. - Using a mouth pipette, collect mitochondria-dense ooplasts from the first bisection drop. Place them in the right-hand T20 drop located in the bottom row of the plate. Repeat for all remaining bisection drops.
NOTE: Mitochondria-dense regions will have a visible conglomeration of organelles that will be light gray in appearance. - Retrieve the T10 four-well dish from the incubator. Using a mouth pipette, move all mitochondria-reduced ooplasts to the well labeled "MR," and move the mitochondria-dense ooplasts to the well labeled "M."
- Place the four-well plate back into the CO2-controlled incubator. Allow ooplasts to rest for at least 30 min prior to quantification of mtDNA.
6. Quantification of mtDNA
- Use a DNA extraction kit designed to extract material from a small sample to extract DNA from individual samples (single oocytes, single mitochondria-dense ooplasts, single mitochondria-depleted ooplasts).
- Use a DNA quantification method of the choice to ensure that DNA was successfully extracted from each sample. If quantitative polymerase chain reaction (qPCR) will not be completed at that time, freeze the extracted DNA in a labeled tube in a -80 °C freezer.
- Perform qPCR
- Refer to Table 2 to determine the volume of each reagent to add to each qPCR tube, along with the primer sequences. These primers are designed to amplify the 12S region of bovine mtDNA.
- Set the initial denaturation time for 10 min, followed by 35 cycles of: 30 s of denaturation at 94 °C, 15 s of annealing at 60 °C, and 15 s of extension at 72 °C. When the reaction is complete, record the cycle threshold (Ct) values.
NOTE: In order to obtain relative mtDNA copy number values, a standard curve must be produced using samples with known quantities of mtDNA copy numbers, that increase exponentially. The cycle threshold values then need to be manipulated using the following formulas to determine relative mtDNA copy numbers. - Obtain the concentration of the DNA using the equation below:
Concentration of DNA = (10(Ct-intercept/slope)) x tested volume - Obtain the copy number using the equation below:
Copy number =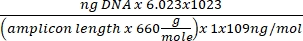
- Obtain the copy number using the equation below:
Copy number per cell = 2 x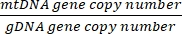
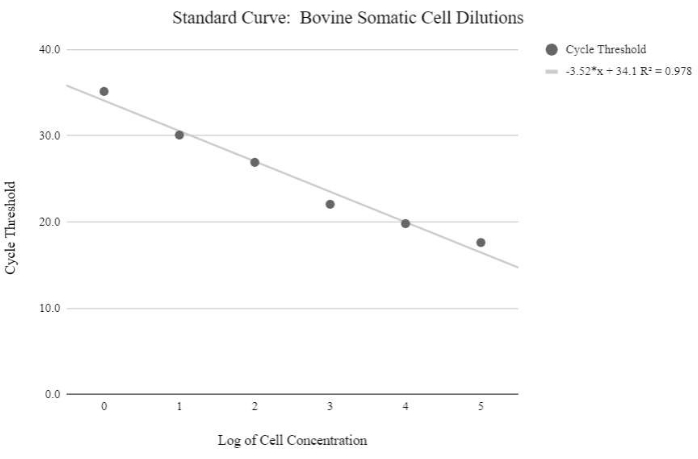
Figure 5: Somatic cell mtDNA standard curve. This standard curve was created through the mtDNA quantification of logarithmic concentrations of bovine somatic cells using the qPCR reagents and program as described in protocol step 6.3. Please click here to view a larger version of this figure.
Representative Results
Quantitative PCR (qPCR) results are used to determine the relative quantities of mtDNA present in each ooplast. The described reaction is designed to amplify the 12S region of bovine mtDNA.
If the bisection was successful, the samples from whole oocytes and mitochondria-dense ooplasts will have similar Ct values. The samples from mitochondria-reduced ooplasts will have higher Ct values when compared to the samples from the other two groups. A Ct graph showing successful bisection results is shown below. These results indicate that bisection did effectively reduce the mtDNA content in the mitochondria-reduced ooplasts (Figure 6).
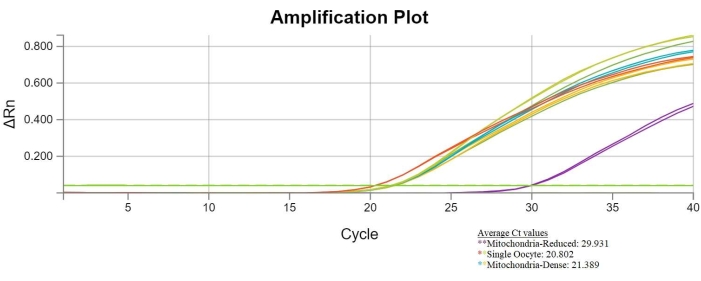
Figure 6: Successful bisection qPCR results. Graph comparing cycle number and fluorescence displays cycle threshold values of single oocytes, mitochondria-reduced ooplasts (represented by purple lines), and mitochondria-dense ooplasts. The Ct values of mitochondria-reduced ooplasts have an average of 29.931 (purple lines), while the average Ct value of the single oocytes is 20.802 (red and green lines), and the average Ct value of the mitochondria-dense ooplasts is 21.389 (blue and gold lines). The increased Ct values of the mitochondria-reduced ooplast samples indicate a decrease in mtDNA copy number within these samples. Please click here to view a larger version of this figure.
A Ct graph showing unsuccessful bisection results is shown in Figure 7. These results indicate that bisection did not effectively reduce the mtDNA content in the mitochondria-reduced ooplasts:
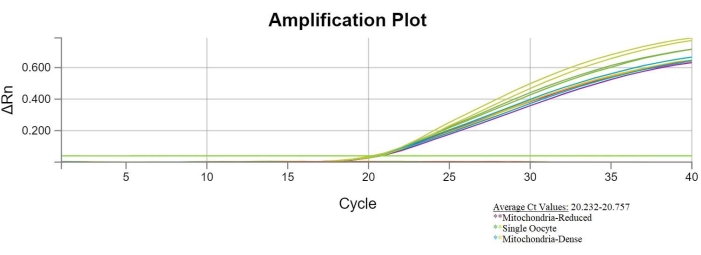
Figure 7: Unsuccessful bisection qPCR results. Line graph comparing cycle number and fluorescence displays cycle threshold values of single oocytes (dark green and light green lines), mitochondria-reduced ooplasts (purple lines), and mitochondria-dense ooplasts (blue and gold lines), all of which have Ct values within the range of 20.232-20.757. This indicates the absence of a significant change in mtDNA copy number in the mitochondria-reduced ooplast samples. Please click here to view a larger version of this figure.
Following the production of a standard curve and use of the provided mtDNA copy number formulas, an average oocyte mtDNA reduction of 93.88% has been achieved using this protocol (Figure 8).
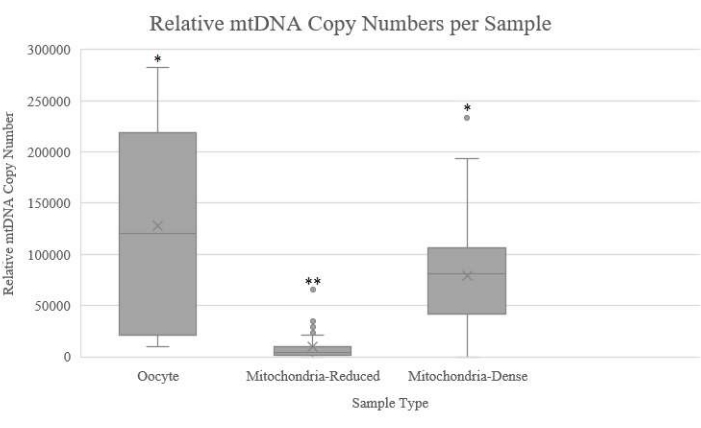
Figure 8: Boxplot comparing relative mitochondrial DNA copy numbers obtained from whole oocytes, mitochondria-reduced ooplasts, and mitochondria-dense ooplasts. Whole oocytes (n = 38), mitochondria-reduced ooplasts (n = 34), and mitochondria-dense ooplasts (n = 29). Mitochondria-reduced ooplasts have significantly fewer copy numbers when compared to the other two groups (P < 0.001). Please click here to view a larger version of this figure.
| Solution | Total Volume | Solvent | Solute |
| Hyaluronidase Solution | 400 µL | 1 mL M-199 | 0.6 mg Hyaluronidase |
| T2 Media | 500 µL | M-199 | 2% Fetal Bovine Serum (v/v) |
| T20 Media | 1020 µL | M-199 | 20% Fetal Bovine Serum (v/v) |
| T10 Media | 800 µL | 400 µL T2 | 400 µL T20 |
| CB/HSOF | 500 µL | 499.5 µL Synthetic Oviductal Fluid with HEPES (HSOF) | 0.5 µL of 10 mg/mL Cytochalasin B (CB) |
| Pronase Solution | 40 µL | 1 mL M-199 | 10 mg pronase |
| T20/CB | 500 µL | 499.5 µL T20 | 0.5 µL of 10 mg/mL CB |
Table 1: Required solutions. Provides volumes of solutes and solvents for each solution required within the protocol.
| Reagent | Volume |
| qPCR master mix | 10 µL |
| Forward primer | 0.6 µL |
| Reverse primer | 0.6 µL |
| DNA sample | 4.4 µL |
| DNase-free water | 4.4 µL |
| Total Volume | 20 µL |
| Forward Primer GGGCTACATTCTCTACACCAAG | |
| Reverse Primer GTGCTTCATGGCCTAATTCAAC | |
Table 2: Quantitative PCR composition. Provides forward and reverse primer sequences and volumes of all reagents needed for each quantitative PCR tube.
Discussion
Methods previously used to decrease mtDNA copy numbers in oocytes have their respective disadvantages. Micromanipulation-based removal of mitochondria from oocytes decrease mtDNA copy numbers by an average of 64%27. A unique method, previously used for enucleation, involves the use of small diameter Pasteur pipettes and the splitting of a zona pellucida-free oocyte at the border between a microdrop of media and the surrounding mineral oil. Along with the utilization of oocyte centrifugation, this approach could produce viable mitochondria-reduced ooplasts28. However, the combination of centrifugation and this oocyte-splitting method has not yet been reported to produce embryos. Chemical mitochondria reduction methods (such as the exposure of oocytes to 2′,3′-dideoxycytidine (ddC), an mtDNA synthesis inhibitor) have reduced oocyte mtDNA copy numbers by up to 50% and require COC's to be exposed to the chemical for over 40 h during in vitro maturation (IVM)16,27. Though incubating porcine oocytes in a ddC-supplemented media for 40 h has not presented issues for the reconstructed embryos16, there is not yet published data regarding the exposure of bovine oocytes to ddC during their 20-22 h IVM or any subsequent embryonic development. The method described in this paper reduces the relative mtDNA copy number by approximately 93.88%, from an average of 45,565 ± 37,169 copies (mean ± SD) to 8,396 ± 13,287.
Several steps within the protocol are critical for obtaining satisfactory results in terms of effectively reducing the mitochondria DNA (mtDNA) copy number of an oocyte. Centrifuging the oocytes at the optimized speed (15,000 x g) and time (12 min) will create a more concentrated mitochondria fraction at one pole of each oocyte. The correct volume of Cytochalasin B/Synthetic Oviductal Fluid with HEPES (CB/HSOF) solution (50 µL) will likely eliminate oocyte lysis due to centrifugation. Following centrifugation, the zona pellucida (ZP) is removed by exposure to a pronase solution; complete removal of the ZP will ensure more precise bisection and should also separate any remaining cumulus cells from the oocytes. The use of a heating stage on the microscope while removing the ZP is optional, but hastens ZP removal by at least 1 min. The orientation of zona pellucida-free (ZF) oocytes in bisection drops is critical to bisecting them accurately. The mitochondria fraction, found at one pole of each oocyte, should be clearly visible and located at either the top or bottom of the oocyte prior to bisection. The opposite pole will contain dark lipid droplets. Following bisection, selection and correct classification of the ooplasts (as mitochondria-dense or mitochondria-depleted) is crucial to obtaining an accurate assessment of the bisection. Selecting which Metaphase II (MII) oocytes to bisect is important as well. Ensuring that the oocytes used are spherical and have homogeneous cytoplasm will likely make a difference in the mtDNA copy numbers contained within each oocyte and ooplast. When designing and synthesizing individual quantitative polymerase chain reactions (qPCR), ensuring that the correct primers and reagent concentrations are used in each reaction is important for obtaining accurate results. Should a standard curve be produced, it should be created as precisely as possible, and all reactions should be performed in triplicate to achieve greater accuracy. The times and temperatures of each qPCR phase should remain the same for every run.
Some issues may arise while following the protocol. When adding oil to the bisection plate, ensure that the drops are just covered; too much oil will make pipette usage less efficient, whereas too little will lead to evaporation of drops. This evaporation would cause a change in the osmolarity of the solutions, which can be harmful to the oocytes and ooplasts. If the oocytes lyse following the digestion of the ZP, this is likely due to one or both of the following, based on personal experience and observation: the oocytes were not centrifuged for the prescribed time and speed, and/or the oocytes were exposed to pronase for too long. Some ZF oocytes may not regain a completely spherical shape within the bisection drops. Keep in mind that not all ZF oocytes will be able to be positioned optimally for bisection. In this case, there are three options: give the ZF oocytes additional time to regain a spherical shape; gently hold oocytes in a more upright position with the microblade prior to bisection; do not bisect oocytes that are unable to be positioned correctly. Determining which ooplasts are mitochondria-dense or mitochondria-depleted after bisection will affect quantification results. If classifying the ooplasts is difficult, position the oocytes that will be bisected so that the mitochondria fraction is either always pointed toward the top or the bottom of the drop, and/or look for a dark portion of the oocyte at one of its poles (these lipid droplets are concentrated at the opposite pole from the mitochondria fraction). At times, ooplasts can stick to one another and/or the indentation made by the microblade in the plate. Sometimes, providing the ooplasts with additional time to regain a spherical shape will assist with separation. Other options include gently tapping the plate, gently sucking them in and out of the pipette, and/or using the edge of the pipette to gently separate the ooplasts.
This mtDNA copy number reduction method does have its limitations; one is based upon oocyte source, and the other is due to the removal of the ZP from the oocyte. Oocyte source (the animal itself, the specific ovary, and the specific follicle) will greatly influence the mtDNA copy numbers within that specific oocyte. There is a large standard deviation around the average mtDNA copy number of a single oocyte, indicating how variable the copy number can be. Many single oocyte quantification reactions are required to have a robust single oocyte standard copy number that will be compared against ooplasts and other oocytes. The sizable standard deviation also makes the comparison of ooplast to oocyte less absolute, as the ooplast's copy number cannot be compared to its oocyte of origin.
The digestion of the oocyte's zonae pellucidae makes the resulting ZF oocyte and ooplasts much more fragile than they were prior to the removal of their zonae pellucidae. This fragility leads to higher lysis rates of ooplasts and the ZF oocytes themselves, particularly with the use of a microblade-a great amount of care and precision must be used. Should the ooplasts be utilized for fusion, they require rest after bisection before fusion can be attempted. Otherwise, many of the ooplasts will likely lyse. The mitochondria-depleted ooplasts tend to be more delicate than their mitochondria-dense counterparts. The microtubules of the oocyte tend to migrate with the mitochondria to the same pole of the oocyte during centrifugation, which leaves the mitochondria-depleted ooplasts with fewer structural components29. Additional research and staining are required to determine if other oocyte organelles are negatively affected by the combination of centrifugation and bisection used to produce ooplasts in this protocol.
The reduction in oocyte mitochondria would reduce the oocyte species' mitochondria in a resulting iSCNT embryo, therefore decreasing the incidence of mitochondria heteroplasmy in that reconstructed embryo. The embryo's chances of development would increase comparatively, but would likely require supplementation of mitochondria from the donor cell species to meet energy requirements during development. Thus far, iSCNT attempts that have used intergeneric crosses have not produced live offspring. Decreasing heteroplasmy in iSCNT embryos may increase the probability of successful pregnancies. iSCNT is a method that could help rescue some of the 40,000 species classified as endangered30. Since somatic cells from these species are typically more accessible than their germ cells, iSCNT can utilize these somatic cells, along with oocytes from domestic species. Intrageneric attempts have been successful in producing live animals31,32,33, though the offspring are typically chimeric for mtDNA, and finding an intrageneric oocyte source is not always possible. Bisection of oocytes with the goal of reducing mtDNA copy numbers, followed by fusion of these ooplasts with an interspecific somatic cell, would reduce chimerism and heteroplasmy in the iSCNT embryos. Because of these reductions, the use of the described method allows for a greater potential of intergeneric live births and the continuation of endangered species.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors wish to thank their colleagues at Utah State University, the Reproductive Science researchers at the San Diego Zoo, and Dr. Rebecca Krisher at Genus PLC.
Materials
| 1.5 mL centrifuge tubes | Fisher Scientific | 5408129 | |
| 60 mm dish | Sigma-Aldrich | D8054 | |
| Centrifuge | Eppendorf | 5424 | |
| Cytochalasin B | Sigma-Aldrich | C6762 | |
| Fetal Bovine Serum | Sigma-Aldrich | F2442 | |
| M199 Media | Sigma-Aldrich | M4530 | |
| Mineral Oil | Sigma-Aldrich | M8410 | |
| Mini Centrifuge | SCILOGEX | D1008 | |
| mtDNA Primer: Forward (12S) | GGGCTACATTCTCTACACCAAG | ||
| mtDNA Primer: Reverse (12S) | GTGCTTCATGGCCTAATTCAAC | ||
| NanoDrop Spectrophotometer | Thermo Scientific | ND2000 | |
| Opthalmic Scalpel with Aluminum Handle | PFM Medical | 207300633 | Microblade for bisection |
| Protease/pronase | Sigma-Aldrich | P5147 | |
| QIAamp DNA Micro Kit | Qiagen | 56304 | |
| QuantStudio™ 3 – 96-Well 0.2-mL | ThermoFisher | A28567 | |
| Search plate | Fisher Scientific | FB0875711A | |
| SYBR Green qPCR Master Mix | ThermoFisher | K0221 | qPCR master mix |
| Synthetic Oviductal Fluid with HEPES (HSOF) | |||
| ThermoPlate | Tokai Hit | TPi-SMZSSX | Heating stage |
References
- Loi, P., Modlinski, J. A., Ptak, G. Interspecies somatic cell nuclear transfer: A salvage tool seeking first aid. Theriogenology. 76 (2), 217-228 (2011).
- Wani, N. A., Vettical, B. S., Hong, S. B. First cloned Bactrian camel (camelus bactrianus) calf produced by interspecies somatic cell nuclear transfer: A step towards preserving the critically endangered wild Bactrian camels. PLOS ONE. 12 (5), 0177800 (2017).
- Oh, H. J., et al. Cloning endangered gray wolves (canis lupus) from somatic cells collected postmortem. Theriogenology. 70 (4), 638-647 (2008).
- Srirattana, K., et al. Full-term development of gaur-bovine interspecies somatic cell nuclear transfer embryos: Effect of trichostatin a treatment. Cellular Reprogramming. 14 (3), 248-257 (2012).
- Kwon, D. K., et al. Blastocysts derived from adult fibroblasts of a rhesus monkey (macaca mulatta) using interspecies somatic cell nuclear transfer. Zygote. 19 (3), 199-204 (2011).
- Lee, E., et al. Production of cloned sei whale (Balaenoptera borealis) embryos by interspecies somatic cell nuclear transfer using enucleated pig oocytes. Journal of Veterinary Science. 10 (4), 285 (2009).
- Lorthongpanich, C., Laowtammathron, C., Chan, A. W., Kedutat-Cairns, M., Parnpai, R. Development of interspecies cloned monkey embryos reconstructed with bovine enucleated oocytes. Journal of Reproduction and Development. 54 (5), 306-313 (2008).
- Hong, S. G., et al. Production of transgenic canine embryos using interspecies somatic cell nuclear transfer. Zygote. 20 (1), 67-72 (2011).
- Stewart, J. B., Chinnery, P. F. The dynamics of mitochondrial DNA heteroplasmy: Implications for human health and disease. Nature Reviews Genetics. 16 (9), 530-542 (2015).
- Takeda, K. Mitochondrial DNA transmission and confounding mitochondrial influences in cloned cattle and pigs. Reproductive Medicine and Biology. 12 (2), 47-55 (2013).
- Lanza, R. P., et al. Cloning of an endangered species (Bos Gaurus) using interspecies nuclear transfer. Cloning. 2 (2), 79-90 (2000).
- Evans, M. J., et al. Mitochondrial DNA genotypes in nuclear transfer-derived cloned sheep. Nature Genetics. 23 (1), 90-93 (1999).
- Meirelles, F. V., et al. Complete replacement of the mitochondrial genotype in a Bos indicus calf reconstructed by nuclear transfer to a Bos taurus oocyte. Genetics. 158 (1), 351-356 (2001).
- Beyhan, Z., Iager, A. E., Cibelli, J. B. Interspecies nuclear transfer: Implications for embryonic stem cell biology. Cell Stem Cell. 1 (5), 502-512 (2007).
- Lagutina, I., Fulka, H., Lazzari, G., Galli, C. Interspecies somatic cell nuclear transfer: advancements and problems. Cellular Reprogramming. 15 (5), 374-384 (2013).
- Jiang, Y., et al. Interspecies somatic cell nuclear transfer is dependent on compatible mitochondrial DNA and reprogramming factors. PLoS ONE. 6 (4), 14805 (2011).
- Chiaratti, M. R., et al. Embryo mitochondrial DNA depletion is reversed during early embryogenesis in cattle. Biology of Reproduction. 82 (1), 76-85 (2010).
- Spikings, E. C., Alderson, J., John, J. C. Regulated mitochondrial DNA replication during oocyte maturation is essential for successful porcine embryonic development. Biology of Reproduction. 76 (2), 327-335 (2007).
- Cagnone, G. L., et al. Restoration of normal embryogenesis by mitochondrial supplementation in pig oocytes exhibiting mitochondrial DNA deficiency. Scientific Reports. 6 (1), 1-15 (2016).
- Spikings, E. C., Alderson, J., John, J. C. Regulated mitochondrial DNA replication during oocyte maturation is essential for successful porcine embryonic development. Biology of Reproduction. 76 (2), 327-335 (2007).
- Ferreira, A. F., et al. Does supplementation with mitochondria improve oocyte competence? A systematic review. Reproduction. 161 (3), 269-287 (2021).
- Bhat, M. H., et al. Live birth of a pashmina goat kid after transfer of handmade cloned embryos. Journal of Reproduction and Development. , (2019).
- Tecirlioglu, R. T., et al. Birth of a cloned calf derived from a vitrified hand-made cloned embryo. Reproduction, Fertility and Development. 15 (7), 361 (2003).
- Zhang, P., et al. Handmade cloned transgenic piglets expressing the nematode fat-1 gene. Cellular Reprogramming. 14 (3), 258-266 (2012).
- Zhang, P., et al. Handmade cloned transgenic sheep rich in omega-3 fatty acids. PLOS ONE. 8 (2), 55941 (2013).
- Lagutina, I., et al. Somatic cell nuclear transfer in horses: Effect of oocyte morphology, embryo reconstruction method and donor cell type. Reproduction. 130 (4), 559-567 (2005).
- Chiaratti, M. R., et al. Embryo mitochondrial DNA depletion is reversed during early embryogenesis in cattle. Biology of Reproduction. 82 (1), 76-85 (2010).
- Hosseini, S. M., et al. and efficient method of manual oocyte enucleation using a pulled pasteur pipette. In Vitro Cellular and Developmental Biology – Animal. 49 (8), 569-575 (2013).
- Zampolla, T., Spikings, E., Rawson, D., Zhang, T. Cytoskeleton proteins F-actin and tubulin distribution and interaction with mitochondria in the granulosa cells surrounding stage III zebrafish (danio rerio) oocytes. Theriogenology. 76 (6), 1110-1119 (2011).
- International Union for Conservation of Nature. The IUCN Red List of Threatened Species. International Union for Conservation of Nature. , (2021).
- Berg, D. K., Li, C., Asher, G., Wells, D. N., Oback, B. Red deer cloned from antler stem cells and their differentiated progeny. Biology of Reproduction. 77 (3), 384-394 (2007).
- Gómez, M. C., et al. Birth of African wildcat cloned kittens born from domestic cats. Cloning and Stem Cells. 6 (3), 247-258 (2004).
- Lanza, R. P., et al. Cloning of an endangered species (Bos gaurus) using interspecies nuclear transfer. Cloning. 2 (2), 79-90 (2000).

