基于RPA-CRISPR/Cas12a-SPM和深度学习的DNA病毒检测系统
Summary
我们提出了一种方案,该方案将重组酶聚合酶扩增与CRISPR / Cas12a系统相结合,用于DNA病毒的痕量检测,并构建了具有人工智能辅助分类的便携式智能手机显微镜,用于即时DNA病毒检测。
Abstract
我们报告了一种快速、易于实施、高度敏感、序列特异性和即时 (POC) DNA 病毒检测系统,该系统结合了重组酶聚合酶扩增 (RPA) 和 CRISPR/Cas12a 系统,用于 DNA 病毒的痕量检测。靶DNA分别被RPA和CRISPR/Cas12a扩增和识别,触发Cas12a的侧支切割活性,切割荧光团淬灭标记的DNA报告基因并泛化荧光。对于POC检测,便携式智能手机显微镜用于拍摄荧光图像。此外,系统内还部署了深度学习模型,用于正样本或负样本的二元分类,实现了高精度。以青蛙病毒3( FV3,Ranavirus属, 虹膜病毒科)为例,对该DNA病毒POC检测系统进行了检测,40 min内检测限(LoD)可达10 aM。无需熟练的操作人员和笨重的仪器,具有人工智能(AI)辅助分类功能的便携式微型RPA-CRISPR/Cas12a-SPM在POC DNA病毒检测方面显示出巨大的潜力,可以帮助防止此类病毒的传播。
Introduction
近年来,由不同病毒引起的传染病流行频发,包括2014年1 和2018年2的埃博拉病毒病(EVD)疫情、2015年中东呼吸综合征(MERS)3、2015年寨卡病毒病(4)疫情、2019年由严重急性呼吸系统综合症冠状病毒2(SARS-CoV-2)5引起的冠状病毒病(COVID-19)和 2022年由猴痘病毒(MKPV)引起的持续猴痘6。这些突如其来的流行性传染病的爆发,造成大量死亡,带来巨大的经济损失和社会动荡。迫切需要一个快速、准确的检测系统,以快速诊断感染并防止病毒的进一步传播。
近年来,成簇规则间隔短回文重复序列(CRISPR)和CRISPR相关(Cas)蛋白引起了全世界的关注,并在核酸检测方面显示出有希望的结果7,8,9,10,11,12,13,14,15 .CRISPR/Cas12a 蛋白在 CRISPR RNA (crRNA) 的指导下,与靶 DNA 结合并切割。这种活性导致非特异性单链DNA(ssDNA)的释放,称为反式裂解,可用于增强核酸检测的检测信号。聚合酶链反应 (PCR)、定量实时 PCR (qPCR) 和酶联免疫吸附测定 (ELISA) 等一些传统检测方法对于即时 (POC) 检测来说既复杂又耗时且成本高昂。我们之前的工作成功开发了一种基于CRISPR/Cas12a技术的非洲猪瘟病毒(ASFV)自动化、集成化和具有成本效益的检测系统。在该系统中,我们在 2 小时的时间内实现了 1 pM 的检测限,无需扩增。将CRISPR/Cas12a系统与重组酶聚合酶扩增(RPA)相结合,提高了痕量DNA检测的灵敏度和特异性。与其他等温扩增技术相比,RPA无需复杂的温度控制设备,反应时间更短,操作方便。
对于病原体的 POC 检测,开发了智能手机显微镜 (SPM)、手持式荧光计或侧向层析条等仪器,用于结果读数 16,17,18。SPM通过摄像头捕捉图像,并将其上传到一些移动应用程序以进行快速数据分析。这种显微镜是一种便携式、廉价和小型化的信号采集系统,具有高灵敏度,在检测 H5N1、寨卡病毒和 SARS-CoV-219,20 等病原体方面显示出优势。因此,我们构建了一个便携式SPM来捕获由RPA-CRISPR / Cas12a检测目标DNA病毒触发的荧光信号。当CRISPR/Cas12a识别到目标DNA病毒时,连接荧光团和淬灭基团的ssDNA报告基因探针将被切割,荧光团发出的荧光可以被SPM捕获。
与通常用于从SPM21的荧光图像中获取结果信息的专业软件相比,一些专家在获得荧光图像22后,使用机器学习和深度学习来量化病毒DNA的浓度,这更加耗时。在对医学图像进行分类时,传统神经网络 (CNN) 通常用于以端到端方式从原始像素化图像中学习特征 23,24,25,26。流行的基于 CNN 的深度学习模型,如 AlexNet、DenseNet-121 和 EfficientNet-B7 已成功应用于该领域27,28。然而,在特定领域获取大型数据集可能具有挑战性,需要迁移学习29,30。该方法使用大型数据集预训练深度学习模型,并将预训练模型用作具有小型数据集的新任务的起点。这种技术可以减少对大型数据集的需求,对抗过拟合,并减少训练时间31。在此,我们使用深度学习模型和迁移学习来对阳性和阴性样本的荧光图像进行二元分类。
在该方法中,我们将RPA和CRISPR / Cas12a系统相结合,用于DNA病毒的痕量检测。靶DNA分别被RPA和CRISPR/Cas12a扩增和识别,触发Cas12a的侧支切割活性,切割荧光团淬灭标记的DNA报告基因并泛化荧光。我们构建了一个便携式SPM来获取荧光图像进行POC检测,并开发用于二元分类的深度学习模型。构建的POC检测系统原理图如图 1所示。在没有熟练的操作人员和笨重的仪器的情况下,具有人工智能(AI)辅助分类的RPA-CRISPR/Cas12a-SPM在POC DNA病毒检测方面显示出巨大的潜力。
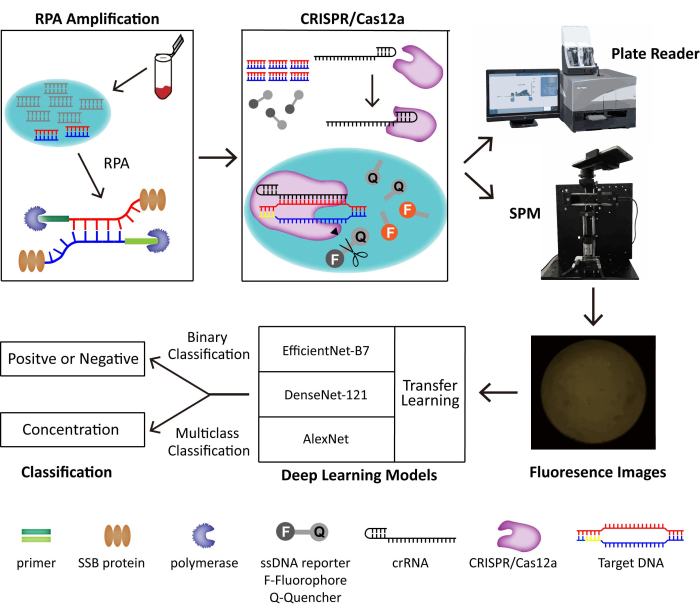
图 1:RPA-CRISPR/Cas12-SPM 检测系统以及用于收集图像的 AI 分类的示意图。 动物源性样本的核酸由PINDBK释放。病毒的靶DNA被RPA-CRISPR/Cas12a系统扩增并特异性识别。CRISPR/Cas12a与crRNA结合,Cas12a-crRNA复合物与靶DNA结合,触发CRISPR/Cas12a在ssDNA报告基因探针上的侧支切割。报告基因上的荧光基团被释放出来,荧光通过商业化的酶标仪或我们制造的 SPM 进行检测。使用三种不同的深度学习模型,包括 AlexNet、DenseNet-121 和具有迁移学习功能的 EfficientNet-B7,对荧光图像进行分类。此图经 Lei et al.35 许可重复使用。 请点击这里查看此图的较大版本.
Protocol
Representative Results
Discussion
在这种方法中,我们开发了一种快速、易于实现、高灵敏度、序列特异性和 POC DNA 病毒检测系统,并具有 AI 辅助。获得样本后,应用RPA对目标序列进行扩增,然后CRISPR/Cas12a可以识别目标DNA并释放荧光,从而放大检测信号。便携式智能手机显微镜用于拍摄荧光图像,具有迁移学习功能的深度学习模型用于正负样本图像的二元分类。该方法以FV3为阳性样本,从多个角度进行研究,以提高所提系统的?…
Disclosures
The authors have nothing to disclose.
Acknowledgements
这项工作得到了国家自然科学基金31970752、深圳市科学技术创新委员会JCYJ20190809180003689、JSGG20200225150707332、JSGG20191129110812708、WDZC20200820173710001的支持;深圳湾实验室开放资助,SZBL2020090501004;中国博士后科学基金 2020M680023;中华人民共和国海关总署 2021HK007。
Materials
| 20x amplification | OLYMPUS | OPLN20X | |
| 532 nm green laser | Thorlabs | PL201 | with 0.9 mW output power |
| 535 nm cutoff wavelength | chrome | AT535 | |
| 6x DNA loading buffer | Thermo scientific | R0611 | |
| 96-well black microplate | Corning Incorporated | 3603 | Black with flat clear bottom |
| Aspherical lens | Lubang | N/A | |
| Bandpass filter | SEMROCK | FF01-542/27-25 | |
| Bsu DNA Polymerase | ATG Biotechnology | M103 | Large Fragment |
| crRNA | Sangon Biotech | N/A | |
| DNA fragments | Sangon Biotech | N/A | |
| Dichroic holders | Ruicage | N/A | |
| Dichroic mirror | SEMROCK | FF555-Di03-25×36 | with a cutoff wavelength of 535 nm |
| E.Z.N.A Gel Extraction Kit | Omega Biotek | D2500-02 | |
| EnGen Lba Cas12a (Cpf1) | New England Biolabs (Beijing) LTD | M0653T | |
| Filter holders | Ruicage | N/A | |
| Fluorophore-ssDNA-Quencher reporter probes | Sangon Biotech | N/A | TAMRA (carboxy tetramethylrhodamine) as the fluorophore at the 5 ends; BHQ2 (Black Hole Quencher-2) as the quencher at the 3 ends |
| GP32 | ATG Biotechnology | M104 | |
| ImageJ | Open-source | Version 1.53t 24 | Downloaded from https://imagej.nih.gov/ij/ |
| Microplate reader | SPARK, TECAN | N/A | |
| Multi-Block thermal Cycler PCR instrument | LongGene | N/A | |
| NanoDrop 2000/2000c Spectrophotometers | Thermo Scientific | ND-2000 | |
| NEBuffer r2.1 | New England Biolabs (Beijing) LTD | B6002S | 10x CRISPR/Cas12a Reaction buffer |
| Oxygen plasma treatment | Electro-Technic Products | N/A | |
| Pathogen Inactivate, Nucleic acid extraction-free, Direct-to-PCR Buffer with Proteinase K (PINDBK) | Ebio | PINDBK -25mL | |
| PCR primer pairs | Sangon Biotech | N/A | |
| PDMS | Dow Corning | Sylgard 184 | |
| RPA primer pairs | Sangon Biotech | N/A | |
| Smartphone | Huawei | Mate10 | |
| Translation stages | Ruicage | N/A | |
| Transmitted neutral density filters | Thorlabs | ND40A | |
| Triplet achromatic lenses | Thorlabs | TRH127-020-A | |
| UvsX | ATG Biotechnology | M105 | |
| UvsY | ATG Biotechnology | M106 |
References
- Gire, S. K., et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 345 (6202), 1369-1372 (2014).
- The Ebola Outbreak Epidemiology Team. Outbreak of Ebola virus disease in the Democratic Republic of the Congo, April-May 2018: an epidemiological study. Lancet. 392 (10143), 213-221 (2018).
- Zumla, A., Hui, D. S., Perlman, S. Middle East respiratory syndrome. Lancet. 386 (9997), 995-1007 (2015).
- Plourde, A. R., Bloch, E. M. A literature review of Zika virus. Emerg Infect Dis. 22 (7), 1185-1192 (2016).
- Yuan, X., et al. Current and perspective diagnostic techniques for COVID-19. ACS Infect Dis. 6 (8), 1998-2016 (2020).
- Minhaj, F. S., et al. Monkeypox outbreak – nine states, May 2022. MMWR Morb Mortal Wkly Rep. 71 (23), 764-769 (2022).
- Bao, M., et al. Challenges and opportunities for clustered regularly interspaced short palindromic repeats based molecular biosensing. ACS Sens. 6 (7), 2497-2522 (2021).
- Broughton, J. P., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 38 (7), 870-874 (2020).
- Chen, J. S., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 360 (6387), 436-439 (2018).
- Gootenberg, J. S., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 356 (6336), 438-442 (2017).
- Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O., Zhang, F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 14 (10), 2986-3012 (2019).
- Mukama, O., et al. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids. Biosens Bioelectron. 159, 112143 (2020).
- Schwank, G., et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 13 (6), 653-658 (2013).
- Yin, L., Man, S., Ye, S., Liu, G., Ma, L. CRISPR-Cas based virus detection: Recent advances and perspectives. Biosens Bioelectron. 193, 113541 (2021).
- Dronina, J., Bubniene, U. S., Ramanavicius, A. The application of DNA polymerases and Cas9 as representative of DNA-modifying enzymes group in DNA sensor design (review). Biosens Bioelectron. 175, 112867 (2021).
- Fozouni, P., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 184 (2), 323-333.e9 (2021).
- Kumar, M., et al. FnCas9-based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip. eLife. 10, e67130 (2021).
- Lee, R. A., et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc Natl Acad Sci U S A. 117 (41), 25722-25731 (2020).
- Ganguli, A., et al. Hands-free smartphone-based diagnostics for simultaneous detection of Zika, Chikungunya, and Dengue at point-of-care. Biomed Microdevices. 19 (4), 73 (2017).
- Yeo, S. J., et al. Smartphone-based fluorescent diagnostic system for highly pathogenic H5N1 viruses. Theranostics. 6 (2), 231-242 (2016).
- von Chamier, L., et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat Commun. 12 (1), 2276 (2021).
- Shiaelis, N., et al. Virus detection and identification in minutes using single-particle imaging and deep learning. ACS Nano. 17 (1), 697-710 (2020).
- Liu, Y., et al. Mixed-UNet: Refined class activation mapping for weakly-supervised semantic segmentation with multi-scale inference. Front. Comput. Sci. 4, 1036934 (2022).
- Lawrimore, J., Doshi, A., Walker, B., Bloom, K. AI-assisted forward modeling of biological structures. Front Cell Dev Biol. 7, 279 (2019).
- Yang, Y., Hu, Y., Zhang, X., Wang, S. Two-stage selective ensemble of CNN via deep tree training for medical image classification. IEEE Trans Cybern. 52 (9), 9194-9207 (2022).
- Zhang, R., et al. RCMNet: A deep learning model assists CAR-T therapy for leukemia. Comput Biol Med. 150, 106084 (2022).
- Xie, Y., et al. Stroke prediction from electrocardiograms by deep neural network. Multimed Tools Appl. 80, 17291-17297 (2021).
- Wang, J., Zhu, H., Wang, S., Zhang, Y. -. D. A review of deep learning on medical image analysis. Mobile Netw Appl. 26, 351-380 (2021).
- Artoni, P., et al. Deep learning of spontaneous arousal fluctuations detects early cholinergic defects across neurodevelopmental mouse models and patients. Proc Natl Acad Sci U S A. 117 (38), 23298-23303 (2020).
- Li, J., et al. DeepLearnMOR: a deep-learning framework for fluorescence image-based classification of organelle morphology. Plant Physiol. 186 (4), 1786-1799 (2021).
- Yosinski, J., Clune, J., Bengio, Y., Lipson, H. How transferable are features in deep neural networks. Proceedings of the 27th International Conference on Neural Information Processing Systems. 2, 3320-3328 (2014).
- He, Q., et al. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. Biosens Bioelectron. 154, 112068 (2020).
- Krizhevsky, A., Sutskever, I., Hinton, G. E. ImageNet classification with deep convolutional neural networks. Commun. ACM. 60 (6), 84-90 (2017).
- Lawton, S., Viriri, S. Detection of COVID-19 from CT lung scans using transfer learning. Comput Intell Neurosci. 2021, 5527923 (2021).
- Lei, Z., et al. Detection of frog virus 3 by integrating RPA-CRISPR/Cas12a-SPM with deep learning. ACS Omega. 8 (36), 32555-32564 (2023).
- Chen, Z., Huang, J., Zhang, F., Zhou, Y., Huang, H. Detection of shrimp hemocyte iridescent virus by recombinase polymerase amplification assay. Mol Cell Probes. 49, 101475 (2020).
- Fu, X., Sun, J., Ye, Y., Zhang, Y., Sun, X. A rapid and ultrasensitive dual detection platform based on Cas12a for simultaneous detection of virulence and resistance genes of drug-resistant Salmonella. Biosens Bioelectron. 195, 113682 (2022).
- Habimana, J. D., et al. Mechanistic insights of CRISPR/Cas nucleases for programmable targeting and early-stage diagnosis: A review. Biosens Bioelectron. 203, 114033 (2022).
- Liang, Y., Lin, H., Zou, L., Deng, X., Tang, S. Rapid detection and tracking of Omicron variant of SARS-CoV-2 using CRISPR-Cas12a-based assay. Biosens Bioelectron. 205, 114098 (2022).
- Sivaraman, D., Biswas, P., Cella, L. N., Yates, M. V., Chen, W. Detecting RNA viruses in living mammalian cells by fluorescence microscopy. Trends Biotechnol. 29 (7), 307-313 (2011).
- Wang, I. H., Burckhardt, C. J., Yakimovich, A., Greber, U. F. Imaging, tracking and computational analyses of virus entry and egress with the cytoskeleton. Viruses. 10 (4), 166 (2018).

