Assay for Blood-brain Barrier Integrity in Drosophila melanogaster
Summary
Blood-brain barrier integrity is critical for nervous system function. In Drosophila melanogaster, the blood-brain barrier is formed by glial cells during late embryogenesis. This protocol describes methods to assay for blood-brain barrier formation and maintenance in D. melanogaster embryos and third instar larvae.
Abstract
Proper nervous system development includes the formation of the blood-brain barrier, the diffusion barrier that tightly regulates access to the nervous system and protects neural tissue from toxins and pathogens. Defects in the formation of this barrier have been correlated with neuropathies, and the breakdown of this barrier has been observed in many neurodegenerative diseases. Therefore, it is critical to identify the genes that regulate the formation and maintenance of the blood-brain barrier to identify potential therapeutic targets. In order to understand the exact roles these genes play in neural development, it is necessary to assay the effects of altered gene expression on the integrity of the blood-brain barrier. Many of the molecules that function in the establishment of the blood-brain barrier have been found to be conserved across eukaryotic species, including the fruit fly, Drosophila melanogaster. Fruit flies have proven to be an excellent model system for examining the molecular mechanisms regulating nervous system development and function. This protocol describes a step-by-step procedure to assay for blood-brain barrier integrity during the embryonic and larval stages of D. melanogaster development.
Introduction
During development, cell-cell communication and interactions are critical for the establishment of tissue and organ structure and function. In some cases, these cell-cell interactions seal off organs from the surrounding environment to ensure proper organ function. This is the case for the nervous system, which is insulated by the blood-brain barrier (BBB). Dysfunction of the BBB in humans has been linked to neurological disorders including epilepsy, and breakdown of the barrier has been observed in neurodegenerative diseases including multiple sclerosis and amyotrophic lateral sclerosis1. In mammals, the BBB is formed by tight junctions between endothelial cells2,3. Other animals, including the fruit fly, Drosophila melanogaster, have a BBB composed of glial cells. These glial cells form a selectively permeable barrier to control movement of nutrients, waste products, toxins, and large molecules into and out of the nervous system4. This allows for the maintenance of the electrochemical gradient necessary to fire action potentials, allowing for mobility and coordination4. In D. melanogaster, the glia protect the nervous system from the potassium-rich, blood-like hemolymph5.
In the central nervous system (CNS) and peripheral nervous system (PNS) of D. melanogaster, two outer glial layers, the subperineurial glia and the perineurial glia, as well as an outer network of extracellular matrix, the neural lamella, form the hemolymph-brain and hemolymph-nerve barrier6, referred to as the BBB throughout this article. During development subperineurial glia become polyploid and enlarge to surround the nervous system5,6,7,8,9,10,11. The subperineurial glia form septate junctions, which provide the main diffusion barrier between the hemolymph and the nervous system5,6,12. These junctions are molecularly similar to the septate-like junctions found at the paranodes of myelinating glia in vertebrates, and they perform the same function as tight junctions in the BBB of mammals13,14,15,16,17. The perineurial glia divide, grow, and wrap around the subperineurial glia to regulate the diffusion of metabolites and large molecules6,10,18,19. BBB formation is complete by 18.5 h after egg laying (AEL) at 25 °C5,8. Previous studies have identified genes that are critical regulators of BBB formation20,21,22. To better understand the exact roles of these genes, it is important to examine the effect of mutation of these potential regulators on BBB integrity. While previous studies have outlined approaches for assaying BBB integrity in embryos and larvae, a comprehensive protocol for this assay has yet to be described5,7. This step-by-step protocol describes methods for assaying BBB integrity during D. melanogaster embryonic and third instar larval stages.
Protocol
1. Collection of Samples
- Embryo collection
- In each embryo collection cage, use a minimum of 50 virgin females with 20−25 males for collections. Incubate these flies in a bottle with cornmeal-agar food (Table of Materials) for 1−2 days before beginning collections23.
NOTE: More flies can be used, but the female-to-male ratio should be kept at 2:1. - Pre-warm apple juice agar plates (Table 1) at 25 °C overnight.
NOTE: This is required for proper staging of embryos. If plates are drying out quickly, add a bowl with water to the incubator to increase the humidity of the chamber. - Anesthetize flies from step 1.1.1 with CO2 and transfer flies to a collection cage. Place a pre-warmed apple juice agar plate with a small smear of yeast paste on the open end and secure to the cage with the red sleeve (Table of Materials). In order to clear older embryos, allow flies to lay embryos/eggs on an apple juice agar plate for 1 h at 25 °C.
- Remove the collection cage from the incubator. Invert the cage mesh side down and tap flies down to the bottom of the cage. Replace the apple juice agar with a new pre-warmed apple juice agar plate with a small smear of yeast paste. Discard the first plate.
- Allow flies to lay embryos/eggs on the new apple juice agar plate for 1 h at 25 °C. Discard this plate following the 1 h collection and proceed to the next step to collect embryos for injection.
- To collect late stage 17 embryos (20−21 h AEL), allow flies of desired genotype from steps 1.1.1−1.1.5 to lay on a new pre-warmed apple juice agar plates with a small smear of yeast paste at 25 °C for 1 h. Age plate for 19 h in a 25 °C incubator, so embryos will be 20−21 h of age at the time of imaging.
NOTE: This step can be repeated as desired for multiple rounds of sample collection, injection, and imaging. - Collect embryos from plates into a cell strainer with 70 μm nylon mesh by adding phosphate-buffered saline (PBS) with 0.1% nonionic surfactant (PBTx; Table 1) to cover the surface of the plate and loosen embryos from the surface using a paintbrush.
- Dechorionate embryos collected in the cell strainer in a 50% bleach solution (Table 1) in a 100 mm Petri dish for 5 min with occasional agitation at room temperature. Rinse embryos 3x by swirling the cell strainer in PBTx in a Petri dish, using a fresh dish of PBTx each time.
- If all embryos are of the correct genotype, proceed directly to step 1.1.10. If generation of embryos of the correct genotype requires a cross with heterozygous flies, select embryos of the correct genotype using the presence or absence of fluorescently marked balancer chromosomes. Use a stereomicroscope with fluorescent capabilities for genotype selection.
NOTE: Balancer chromosomes marked with Deformed-yellow fluorescent protein; Kruppel-Gal4, UAS-green fluorescent protein (GFP); and twist-Gal4, UAS-GFP work well for genotype selection in late embryogenesis (Table 2)24,25,26. - Using a glass pipette, transfer embryos in PBTx to a 2% agarose gel slab (Table 1). Remove excess liquid with filter paper. Align ~6−8 embryos on the 2% agarose gel slab with posterior to the right and dorsal side facing up (Figure 1B).
NOTE: The micropyle, the small hole through which spermatozoa enter the egg, is located at the anterior end of the embryo. The posterior end is more rounded. The trachea appears white and is located dorsally in the embryo, allowing for the distinction of the dorsal and ventral sides of the embryo (Figure 1B). - Prepare a slide with one piece of double-sided tape. Firmly press the slide on top of the embryos to transfer them to the double-sided tape.
- Desiccate embryos by incubation at room temperature for ~25 min (no desiccant is used). Following desiccation, cover embryos with halocarbon oil.
NOTE: Desiccation periods may vary depending on the temperature, humidity, and ventilation in the room. The incubation period should be used to set up the apparatus for injection and the confocal microscope for imaging, as described in sections 2 and 3 of this protocol.
- In each embryo collection cage, use a minimum of 50 virgin females with 20−25 males for collections. Incubate these flies in a bottle with cornmeal-agar food (Table of Materials) for 1−2 days before beginning collections23.
- Larval collection
- Set up a cross with 5−10 virgin female flies of the desired genotype and half as many males of the desired genotype in a vial with cornmeal-agar food and incubate at 25 °C23.
- After 5−7 days, depending on the genotype, collect wandering third instar larvae from the vial gently with forceps. Rinse larvae in 1x PBS to remove food stuck to the larvae. Transfer larvae to an apple juice agar plate for genotyping as described in step 1.1.9 if necessary.
- Roll larvae on a tissue using a paintbrush to dry them off. Transfer 6−8 larvae to a slide prepared with double-sided tape using forceps.
2. Preparation of Needles and Specimen Injection
- Pull needles on a micropipette puller prior to the initiation of this protocol. Secure capillary tubes into the needle puller and pull according to the standard needle shape and parameters for D. melanogaster injections (Table 3)27. Store needles in a Petri dish by anchoring in clay until use for injection.
- Load a needle with 5 μL of 10 kDa dextran conjugated to sulforhodamine 101 acid chloride (Table of Materials) using a 20 μL gel-loading pipette tip during the 25-min desiccation period for embryos (step 1.1.12), or immediately following the transfer of larvae to the slide (step 1.2.3).
- Load the needle into a needle holder and position in a micromanipulator secured to a steel base (Table of Materials).
NOTE: The needle should be nearly parallel to the microscope stage for embryo injection and angled slightly downward for larval injections. - Set the injection apparatus (Table of Materials) to 50 psi, 5−10 ms with a range of 10.
NOTE: It may be necessary to alter these settings for the particular injection apparatus being used. - Place the slide on stage and brush edge of the needle against the edge of the double-sided tape at a 45° angle to create an angled, broken tip.
NOTE: For embryos it is only necessary to break the tip enough to allow for flow of the 10 kDa dextran. A perfect needle has a slightly angled tip and only a small drop of dye will come out with each injection. For larvae, it is necessary to break the tip more, but with an angled tip to penetrate the larval body wall. A larger drop of dye will come out. - Pump foot pedal until the dye is at the tip of the needle.
- Align the needle so that it is parallel with the embryo or angled slightly downward toward the larva.
- Move the needle to puncture the posterior end of the specimen and inject the specimen by pumping the foot pedal. Inject ~2 nL of dye into embryos, and ~220 nL of dye into larvae.
NOTE: The embryo or larva should flood with dye if the injection is successful. - Note the time of injection for incubation purposes. Incubate embryos for 10 min at room temperature. Incubate larvae for 30 min at room temperature.
- Continue down the slide to inject additional specimens, noting the time of injection for each specimen.
NOTE: Depending on the speed with which subsequent dissection and imaging steps can be performed, 4−8 specimens can be injected at a time.
3. Preparation of Samples for Imaging
- Imaging of embryos
- Following injection, prepare embryos for imaging. Apply petroleum jelly with a cotton-tipped applicator on the right and left sides of the samples on the slide as a spacer to prevent damage to the embryos upon placement of the coverslip.
- Image samples using a confocal microscope throughout the depth of the embryo with a 20x objective. Calculate the percentage of total samples with dye observed in the ventral nerve cord (VNC) using the following equation: % of samples with compromised BBB = Number of samples with dye accumulation in the VNC/total number of samples assayed.
- Dissection and imaging of larvae
- Prepare slides for larval samples ahead of time. Mount two coverslips spaced approximately 0.5 cm apart to the slide with nail polish.
NOTE: The coverslips function as spacers for the brain, so it is not damaged during the mounting process. - Following the 30 min incubation, dissect the larvae in 1x PBS directly on the slide that will be used in imaging. First, use one pair of forceps to grab the larva halfway down the larval body, and use a second pair of forceps to separate the anterior and posterior halves of the larva.
- Next, use one pair of forceps to grip the anterior region at the mouth hooks, and use a second pair of forceps to invert the body wall over the tip of the forceps gripping the mouth hooks. The brain and VNC will be exposed.
- Separate the brain and VNC from the body wall by severing the nerves, and remove the body wall from the slide (Figure 1C,D). Remove imaginal discs if desired.
- Cover the sample with 10 μL of 80% glycerol and place a coverslip on top of the sample for imaging.
- Image through the depth of the nervous system tissue using a 20x objective. Calculate the percentage of total samples with dye observed in the VNC.
- Prepare slides for larval samples ahead of time. Mount two coverslips spaced approximately 0.5 cm apart to the slide with nail polish.
Representative Results
The methods described here allow for the visualization of the integrity of the BBB throughout the CNS in D. melanogaster embryos and larvae (Figure 1). Upon completion of BBB formation in late embryogenesis, the BBB functions to exclude large molecules from the brain and VNC5. This protocol takes advantage of this function to assay BBB formation. When wild-type (Oregon R) late stage 17 (20−21 h old) embryos were injected with 10 kDa dextran conjugated to sulforhodamine 101 acid chloride fluorescent dye, the large dextran molecule was excluded from the VNC, as expected (Figure 2A). In order to demonstrate the effect of genetic mutations on BBB integrity, embryos with mutations in the raw gene were utilized. Previously, the raw gene has been demonstrated to regulate germ band retraction during embryogenesis, and more recently has been shown to function in glia to regulate morphological changes in the VNC during development28. Heterozygous raw1 mutant embryos exhibited an intact BBB, similar to the results observed in wild-type control embryos (Figure 2B). In contrast, homozygous raw1 mutant embryos exhibited defects in the integrity of the BBB, with 10 kDa dextran dye flooding into the VNC, indicating that the BBB failed to form (Figure 2C). These results demonstrate the ability of this technique to assay BBB formation during embryonic stages.
Previous studies have demonstrated that a defect in subperineurial glia polyploidization results in a disruption of the BBB that can be observed in third instar larval stages7,9. Thus, defects in BBB formation and/or maintenance could result in a compromised BBB during later stages of development, making it desirable to assay integrity of the barrier in the third instar larval stage. Therefore, the protocol utilized for assaying BBB integrity in embryonic stages has also been optimized for use in larvae. In Oregon R control samples, 10 kDa dextran fails to penetrate the BBB and is excluded from the brain and VNC (Figure 2D,E). The dye accumulates at the periphery of the BBB.
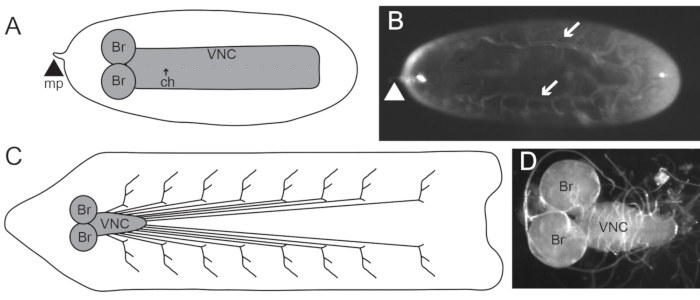
Figure 1: The nervous system of stage 17 embryos and third instar larva. (A) Schematic of a ventral view of a stage 17 embryonic central nervous system (CNS). The CNS consists of the brain (Br) and ventral nerve cord (VNC), which has dorsoventral channels (ch). The micropyle (mp; arrowhead) at the anterior end can be used to orient the embryo. Posterior to the right. (B) Stage 17 embryo oriented with the dorsal side up and posterior to the right, as recommended in step 1.1.10. Arrows are pointing to the trachea. Arrowhead indicates the mp. Posterior to the right. (C) Schematic of the nervous system in the third instar larva. The brain (Br) and VNC compose the CNS, while the nerves extending from the VNC synapse onto body wall muscles and are part of the PNS. Posterior to the right. (D) Dissected third instar larval brain and VNC. Posterior to the right. Please click here to view a larger version of this figure.
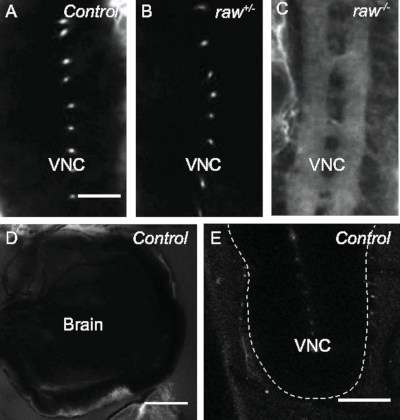
Figure 2: Assay for blood-brain barrier (BBB) formation. (A-C) Late stage 17 embryos (20−21 h old) injected posteriorly with 10 kDa sulforhodamine 101 acid chloride dextran. Posterior down. Scale bar = 20 μm. Dots seen in controls are dorsoventral channels that span the ventral nerve cord (VNC). (A) Oregon R control. Dye uptake in 6.25% of samples, n = 16. (B) raw1/+ sibling control. Dye uptake in 6.67% of samples, n = 15. (C) Homozygous raw1/1 mutant. Dye uptake in 100% of samples, n = 22. (D) Oregon R control third instar larval brain. Dye accumulates at the BBB, but does not penetrate into the CNS, n = 7. Scale bar = 50 μm. (E) Oregon R control third instar larval VNC. Dye accumulates at the BBB, but does not penetrate into the CNS, n = 11. Dashed line outlines the VNC. Scale bar = 50 μm. Please click here to view a larger version of this figure.
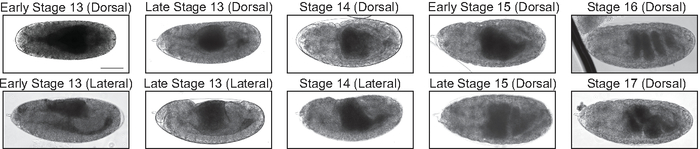
Figure 3: Midgut morphology in late embryonic development. Transmitted light images of stage 13−17 embryos allow for visualization of gut morphology (dark grey regions in the posterior half of the embryo). Midgut morphology can be used to determine the stage of embryonic development, which is helpful when determining if embryos are being aged appropriately for injection. Scale bar = 100 μm. Please click here to view a larger version of this figure.
| Solution | Recipe |
| 2% Agarose gel | Add 0.8 g of agarose to 40 mL of double-distilled H2O (ddH2O) in an Erlenmeyer flask and microwave to dissolve agar. Pour into a gel casting tray and allow the gel to solidify for 30 min at room temperature. |
| Apple juice agar plates | Measure 45 g of agar and 1.5 L of ddH2O into a 4 L flask. Autoclave for 40−45 min at 121 °C. Measure 50 g of sugar and 0.5 L of apple juice into a 1 L beaker and stir on low heat (setting 3) to dissolve sugar, taking care not to burn it. Following autoclaving, add the preheated sugar and apple juice to the agar and water. Stir on low with the heat off to allow to cool until you can touch it. Add 15 mL of 70% tegosept and stir to disperse. Pour into a 0.5 L beaker. Spray with ethanol to remove bubbles or flame with a Bunsen burner. Pour into 60 mm petri dishes. Allow to set for at least 24 h, or until there is minimal condensation on the lids of the petri dishes and store at 4 °C. |
| 50% Bleach | Add 15 mL of ddH2O and 15 mL of household bleach into a conical tube. |
| 80% Glycerol | For 10 mL, add 8 mL of autoclaved ddH2O and 2 mL of glycerol to a 15 mL conical tube. Incubate on a rocker until solution is homogeneous. |
| 1x PBS | For 1 L, dilute 100 mL of 10x PBS in 900 mL of ddH2O. |
| 10x PBS | For 2 L, dissolve the following in 1,600 mL of ddH2O: 160 g of NaCl, 4 g of KCl, 28.8 g of Na2HPO4, and 4.8 g of KH2PO4. Adjust pH to 7.5 with HCl. |
| PBTx (PBS + 0.1% nonionic surfactant) | For 1 L, combine 100 mL of 10x PBS, 10 mL of 10% nonionic surfactant, and 890 mL of ddH2O. |
| 70% Tegosept | Mix 1 g of p-hydroxybenzoic acid, methyl ester for every 10 mL of 100% ethanol. Store at -20 °C. |
| 10% Nonionic surfactant | For 50 mL, add 45 mL of autoclaved ddH2O and 5 mL of nonionic surfactant to a conical tube. Rotate on the rocker until the solution is homogeneous. |
| Yeast paste | Mix dry active yeast with ddH2O in a 50 mL plastic beaker until smooth. |
Table 1: Reagents and buffers used throughout this protocol.
| Genotype | Stock |
| w[1118]; ln(2LR)Gla, wg[Gla-1]/CyO, P{w[+mC]=GAL4-twi.G}2.2, P{w[+mC]=UAS-2xEGFP}AH2.2 | Bloomington #6662 |
| w{*]; sna[Sco]/CyO, P{w[+mC]=Dfd-EYFP}2 | Bloomington #8578 |
| w{*]; L[2] Pin[1]/CyO, P{w[+mC]=GAL4-Kr.C}DC3, P{w[+mC]=UAS=GFP.S65T}DC7 | Bloomington #5194 |
| Df(1)JA27/FM7c, P{w[+mC]=GAL4-Kr.C}DC1, P{w[+mC]=UAS-GFP.S65T}DC5, sn[+] | Bloomington #5193 |
| w[*]; ry[506] Dr[1]/TM6B, P{w[+mc]=Dfd-EYFP}3, Sb[1] Tb[1] ca[1] | Bloomington #8704 |
| y[1] w[*]; D[*] gl[3]/TM3, P{w[+mC]=GAL4-Kr.C}DC2, P{w[+mC]=UAS-GFP.S65T}DC10, Sb[1] | Bloomington #5195 |
| Oregon R | Multiple Strains Available |
| raw[1] cn[1] bw[1] sp[1]/CyO | Bloomington #2749 |
| P{w[+mW.hs]=GawB}elav[C155] | Bloomington #458 |
| w[1118]; P{w[+m*]=GAL4}repo/TM3, Sb[1] | Bloomington #7415 |
| P{UAS-GFP} | Multiple strains available |
Table 2: Fly strains. Fly strains discussed throughout this protocol. The Bloomington Drosophila Stock Center stock number is provided where applicable.
| Cycle | Heat | Pull | Velocity | Time | Pressure | Ramp |
| 1 | 590 | 115 | 15 | 250 | 600 | X |
| 2 | 575 | 130 | 60 | 250 | 600 | X |
Table 3: Micropipette puller settings. Micropipette puller settings used to generate needles for injection in this protocol.
Discussion
This protocol provides a comprehensive description of the steps needed to assay for BBB integrity during the late embryonic and third instar larval stages of D. melanogaster development. Similar approaches have been described elsewhere to assay the integrity of the BBB during development, as well as in adult stages5,7,29,30. However, descriptions of procedures in materials and methods sections are often broad in nature and lack sufficient detail for easy implementation, necessitating significant troubleshooting on behalf of the researcher. This protocol provides a comprehensive description of the steps needed to assay for BBB integrity during embryonic and larval stages. In each stage, there are critical steps to be followed to ensure that the correct stage of development is being examined and tissue architecture is not disrupted, which could compromise the BBB. In order to achieve success in these approaches it was necessary to troubleshoot multiple steps of the protocol so as to yield accurate results. Critical steps in the embryonic and larval protocols are described below.
Previous studies have reported the establishment of the BBB by 18.5 h AEL5. To ensure accurate developmental timing, it is important to maintain samples at a constant temperature during aging. When collecting embryos, apple juice agar plates should be brought to 25 °C prior to use for collection. Using apple juice plates that have not been pre-warmed results in slower development and can yield samples that have not yet established a BBB and would therefore exhibit accumulation of 10 kDa dextran in the nervous system. As such, it is important to make sure that one is injecting samples that are older than 18.5 h. Additionally, when performing these experiments in embryos, it is necessary to consider the possibility of a developmental delay, because such a delay could simply result in the later establishment of the BBB. To account for any potential developmental delays, samples were assayed at 20−21 h AEL, slightly after reported BBB formation. Using the morphology of other tissues, in particular the developing midgut, can assist in establishing the correct stage of development of the embryo being assayed (Figure 3). Conversely, improper experimental timing can result in samples that are already motile and have begun the hatching process. To reduce the number of larvae that have already started hatching, it is important to perform a minimum of two 1 h collections to clear older embryos from females before collecting embryos for injection. If analysis of the BBB in first instar larvae is desired, a brief incubation of larvae in 1x PBS in a 9-well dish on ice can be used to reduce motility prior to injection. Slides can also be kept on an ice pack during the injection procedure to reduce motility.
In order to ensure quality images and accurate results regarding the establishment of the BBB in the embryo, additional steps must be carefully followed throughout the procedure. When lining up embryos on the agar slab, the trachea should be facing up, as this is the dorsal side of the embryo (Figure 1B). When the embryos are transferred to the slide with the double-sided tape, the ventral side of the embryo will then be facing up, allowing for optimal visualization of the nerve cord during confocal imaging later in the protocol. Embryos must also be securely adhered to the double-sided tape to inhibit movement during injection. The use of a flat 2% agarose pad allows the researcher to push the slide with the double-sided tape firmly onto the embryos during the transfer process without risk of damage to the embryos. Following transfer to the slide, desiccation is necessary to prevent the embryo from bursting upon dye injection. Injection of too much dye can also cause the embryo to burst. It is important to only insert the needle into the embryo enough to inject the dye, but not so much that the needle potentially punctures the nervous system, which would give a false positive result. Too large of a puncture wound can also result in the hemolymph and dye flooding out of the embryo, leading to a failed experiment. With these potential pitfalls in mind, injection is most successful with a needle that has a slight angle to the tip, such that there is a small point with which to carefully puncture the embryo.
In the case of assaying larvae for BBB integrity, challenges can arise due to the motility of the sample. The use of high-quality double-sided tape is critical, as it should be effective in immobilizing larvae for injection. If the larva does become unstuck, it can be rolled back on the tape and gently pushed down to resecure it. When transporting larvae for injection, it is helpful to carry the slide in a Petri dish in case the larvae become unstuck. The steps following injection require the most care to avoid disruption of the BBB. In order to minimize potential damage to the sample, it is easiest to dissect the sample directly on the slides that will be used for imaging. If the larva is strongly adhered to the tape when transferring the sample for dissection, a drop of 1x PBS can be added to the tape to loosen adhesion and to allow for transfer to the slide for dissection. Following dissection, it is important to flatten the sample sufficiently to avoid false positive results, but not so much that sample integrity is compromised. Sandwiching the larva between the coverslip and the slide using additional coverslips as spacers prevents the brain from being damaged, yet immobilizes the sample for effective imaging.
In order to achieve success with protocols for both embryos and larvae, it is critical to make sure the injection apparatus and confocal microscope are set up before sample processing begins. This allows for accurate timing in the imaging of samples, which is necessary for sample comparison. Other approaches have utilized more advanced injection set ups, requiring an inverted microscope set up for embryo injection. This protocol utilizes a standard dissecting microscope and micromanipulator with a pressure regulator. In this case, a zebrafish injection set up was simply adapted for injection of fly embryos and larvae. This protocol could be simplified further to utilize a micromanipulator with a syringe for injection as well. Many of these tools are readily available in Biology departments and may even be available in classroom laboratory settings, allowing for maximal ease of protocol implementation.
During the imaging procedure, it can be helpful to label the cells of the nervous system to more easily identify the desired region for imaging. Specifically, one could use the GAL4/UAS system to express GFP in either neurons or glia, using the elav-Gal4 or repo-Gal4 strains, respectively (Table 2)31,32,33. Such labeling would provide a contrast with the sulforhodamine 101 acid chloride dextran to visualize the integrity of the nervous system.
While this method is focused on assaying the integrity of the BBB during development of D. melanogaster, this approach could be adapted to examine the integrity of other barriers across a variety of organisms and tissues. For example, similar protocols have been published for assaying the BBB in mice34. In addition, permeability of the blood-eye barrier and the somatic barrier surrounding the germ cells during spermatogenesis in D. melanogaster utilize a similar approach29,35. The protocol could also be adapted for use in other tissues to test permeability, including in the intestine. When adapting this protocol for use in other tissues or species, it will be necessary to consider the size of molecules that can penetrate the barrier, as it is feasible that 10 kDa dextran may be small enough to permeate some barriers. Overall, this protocol provides a step-by-step procedure to assay for BBB integrity during D. melanogaster development that can be easily adapted for use in other settings.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The authors thank Dr. F. Bryan Pickett and Dr. Rodney Dale for use of equipment for injection. This work was funded by research funding from Loyola University Chicago to M.D., D.T., and J.J.
Materials
| 10 kDa sulforhodamine 101 acid chloride (Texas Red) Dextran | ThermoFisher Scientific | D1863 | Dextran should be diluted in autoclaved ddH2O to a concentration of 25 mg/mL. |
| 20 μL Gel-Loading Pipette Tips | Eppendorf | 22351656 | |
| 100% Ethanol (200 proof) | Pharmco-Aaper | 11000200 | |
| Active Dry Yeast | Red Star | ||
| Agar | Fisher Scientific | BP1423 | |
| Agarose | Fisher Scientific | BP160-500 | |
| Air Compressor | DeWalt | D55140 | |
| Apple Juice | Mott's Natural Apple Juice | ||
| Bleach | Household Bleach | 1-5% Hypochlorite | |
| Borosilicate Glass Capillaries | World Precision Instruments | 1B100F-4 | |
| Bottle Plugs | Fisher Scientific | AS-277 | |
| Cell Strainers | BD Falcon | 352350 | |
| Confocal Microscope | Olympus | FV1000 | Samples imaged using 20x objective (UPlanSApo 20x/ 0.75) |
| Cotton-Tipped Applicator | Puritan | 19-062614 | |
| Double-Sided Tape 1/2" | Scotch | ||
| Dumont Tweezers; Pattern #5; .05 X .01mm Tip | Roboz | RS-5015 | |
| Fly Food Bottles | Fisher Scientific | AS-355 | |
| Fly Food Vials | Fisher Scientific | AS-515 | |
| Foot Pedal | Treadlite II | T-91-S | |
| Gel Caster | Bio-Rad | 1704422 | |
| Gel Tray | Bio-Rad | 1704436 | |
| Glass Pipette | VWR | 14673-010 | |
| Glycerol | Fisher Scientific | BP229-1 | |
| Granulated sugar | Purchased from grocery store. | ||
| Halocarbon Oil | Lab Scientific, Inc. | FLY-7000 | |
| Light Source | Schott | Ace I | |
| Manipulator Stand | World Precision Instruments | M10 | |
| Micromanipulator | World Precision Instruments | KITE-R | |
| Micropipette Puller | Sutter Instrument Co. | P-97 | |
| Needle Holder | World Precision Instruments | MPH310 | |
| Nightsea Filter Sets | Electron Microscopy Science | SFA-LFS-CY | For visualization of YFP |
| Nightsea Full Adapter System w/ Royal Blue Color Light Head | Electron Microscopy Science | SFA-RB | For visualization of GFP |
| Paintbrush | Simply Simmons | Chisel Blender #6 | |
| Pipetter | Fisher Scientific | 13-683C | |
| Pneumatic Pump | World Precision Instruments | PV830 | This is also referred to as a microinjector or pressure regulator. Since the model used in our study is no longer available this is one alternative. |
| Potassium Chloride | Fisher Scientific | BP366-500 | |
| Potassium Phosphate Dibasic | Fisher Scientific | BP363-500 | |
| Small Embryo Collection Cages | Genesee Scientific | 59-100 | |
| Sodium Chloride | Fisher Scientific | BP358-212 | |
| Sodium Phosphate Dibasic Anhydrous | Fisher Scientific | BP332-500 | |
| Steel Base Plate | World Precision Instruments | 5052 | |
| Stereomicroscope | Carl Zeiss | Stemi 2000 | Used for tissue dissection. |
| Stereomicroscope with transmitted light source | Baytronix | Used for injection. | |
| Tegosept (p-hydroxybenzoic acid, methyl ester) | Genesee Scientific | 20-258 | |
| Triton X-100 | Fisher Scientific | BP151-500 | Nonionic surfactant |
| Vial Plugs | Fisher Scientific | AS-273 |
Referenzen
- Obermeier, B., Daneman, R., Ransohoff, R. M. Development, maintenance and disruption of the blood-brain barrier. Nature Medicine. 19 (12), 1584-1596 (2013).
- Brightman, M. W., Reese, T. S. Junctions between intimately apposed cell membranes in the vertebrate brain. Journal of Cell Biology. 40 (3), 648-677 (1969).
- Tietz, S., Engelhardt, B. Brain barriers: Crosstalk between complex tight junctions and adherens junctions. Journal of Cell Biology. 209 (4), 493-506 (2015).
- Hindle, S. J., Bainton, R. J. Barrier mechanisms in the Drosophila blood-brain barrier. Frontiers in Neuroscience. 8, 414 (2014).
- Schwabe, T., Bainton, R. J., Fetter, R. D., Heberlein, U., Gaul, U. GPCR signaling is required for blood-brain barrier formation in drosophila. Cell. 123 (1), 133-144 (2005).
- Stork, T., et al. Organization and function of the blood-brain barrier in Drosophila. Journal of Neuroscience. 28 (3), 587-597 (2008).
- Unhavaithaya, Y., Orr-Weaver, T. L. Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes & Development. 26 (1), 31-36 (2012).
- Schwabe, T., Li, X., Gaul, U. Dynamic analysis of the mesenchymal-epithelial transition of blood-brain barrier forming glia in Drosophila. Biology Open. 6 (2), 232-243 (2017).
- Von Stetina, J. R., Frawley, L. E., Unhavaithaya, Y., Orr-Weaver, T. L. Variant cell cycles regulated by Notch signaling control cell size and ensure a functional blood-brain barrier. Development. 145 (3), dev157115 (2018).
- von Hilchen, C. M., Beckervordersandforth, R. M., Rickert, C., Technau, G. M., Altenhein, B. Identity, origin, and migration of peripheral glial cells in the Drosophila embryo. Mechanisms of Development. 125 (3-4), 337-352 (2008).
- Beckervordersandforth, R. M., Rickert, C., Altenhein, B., Technau, G. M. Subtypes of glial cells in the Drosophila embryonic ventral nerve cord as related to lineage and gene expression. Mechanisms of Development. 125 (5-6), 542-557 (2008).
- Bellen, H. J., Lu, Y., Beckstead, R., Bhat, M. A. Neurexin IV, caspr and paranodin–novel members of the neurexin family: encounters of axons and glia. Trends in Neurosciences. 21 (10), 444-449 (1998).
- Baumgartner, S., et al. A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell. 87 (6), 1059-1068 (1996).
- Banerjee, S., Pillai, A. M., Paik, R., Li, J., Bhat, M. A. Axonal ensheathment and septate junction formation in the peripheral nervous system of Drosophila. Journal of Neuroscience. 26 (12), 3319-3329 (2006).
- Bhat, M. A., et al. Axon-glia interactions and the domain organization of myelinated axons requires neurexin IV/Caspr/Paranodin. Neuron. 30 (2), 369-383 (2001).
- Faivre-Sarrailh, C., et al. Drosophila contactin, a homolog of vertebrate contactin, is required for septate junction organization and paracellular barrier function. Development. 131 (20), 4931-4942 (2004).
- Salzer, J. L., Brophy, P. J., Peles, E. Molecular domains of myelinated axons in the peripheral nervous system. Glia. 56 (14), 1532-1540 (2008).
- von Hilchen, C. M., Bustos, A. E., Giangrande, A., Technau, G. M., Altenhein, B. Predetermined embryonic glial cells form the distinct glial sheaths of the Drosophila peripheral nervous system. Development. 140 (17), 3657-3668 (2013).
- Matzat, T., et al. Axonal wrapping in the Drosophila PNS is controlled by glia-derived neuregulin homolog Vein. Development. 142 (7), 1336-1345 (2015).
- Limmer, S., Weiler, A., Volkenhoff, A., Babatz, F., Klambt, C. The Drosophila blood-brain barrier: development and function of a glial endothelium. Frontiers in Neuroscience. 8, 365 (2014).
- Ho, T. Y., et al. Expressional Profiling of Carpet Glia in the Developing Drosophila Eye Reveals Its Molecular Signature of Morphology Regulators. Frontiers in Neuroscience. 13, 244 (2019).
- DeSalvo, M. K., et al. The Drosophila surface glia transcriptome: evolutionary conserved blood-brain barrier processes. Frontiers in Neuroscience. 8, 346 (2014).
- . BDSC Cornmeal Food Available from: https://bdsc.indiana.edu/information/recipes/bloomfood.html (2017)
- Le, T., et al. A new family of Drosophila balancer chromosomes with a w- dfd-GMR yellow fluorescent protein marker. Genetik. 174 (4), 2255-2257 (2006).
- Casso, D., Ramirez-Weber, F. A., Kornberg, T. B. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mechanisms of Development. 88 (2), 229-232 (1999).
- Halfon, M. S., et al. New fluorescent protein reporters for use with the Drosophila Gal4 expression system and for vital detection of balancer chromosomes. Genesis. 34 (1-2), 135-138 (2002).
- Miller, D. F., Holtzman, S. L., Kaufman, T. C. Customized microinjection glass capillary needles for P-element transformations in Drosophila melanogaster. BioTechniques. 33 (2), 366-372 (2002).
- Luong, D., Perez, L., Jemc, J. C. Identification of raw as a regulator of glial development. PLoS One. 13 (5), e0198161 (2018).
- Pinsonneault, R. L., Mayer, N., Mayer, F., Tegegn, N., Bainton, R. J. Novel models for studying the blood-brain and blood-eye barriers in Drosophila. Methods in Molecular Biology. 686, 357-369 (2011).
- Love, C. R., Dauwalder, B., Barichello, T. Drosophila as a Model to Study the Blood-Brain Barrier. Blood-Brain Barrier. , 175-185 (2019).
- Lin, D. M., Goodman, C. S. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 13 (3), 507-523 (1994).
- Sepp, K. J., Schulte, J., Auld, V. J. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Entwicklungsbiologie. 238 (1), 47-63 (2001).
- Brand, A. H., Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118 (2), 401-415 (1993).
- Devraj, K., Guerit, S., Macas, J., Reiss, Y. An In Vivo Blood-brain Barrier Permeability Assay in Mice Using Fluorescently Labeled Tracers. Journal of Visualized Experiments. 132, e57038 (2018).
- Fairchild, M. J., Smendziuk, C. M., Tanentzapf, G. A somatic permeability barrier around the germline is essential for Drosophila spermatogenesis. Development. 142 (2), 268-281 (2015).

