用于微生物化学和蛋白质生产的光控发酵
Summary
微生物代谢的光遗传学控制为发酵过程提供了灵活的动态控制。这里的实验方案展示了如何建立蓝光调节的发酵,用于不同体积尺度的化学和蛋白质生产。
Abstract
微生物细胞工厂为从可再生原料生产化学品和重组蛋白提供了一种可持续的替代方案。然而,通过基因修饰使微生物负担过重会降低宿主的适应性和生产力。这个问题可以通过使用动态控制来克服:酶和途径的诱导表达,通常使用基于化学或营养的添加剂,以平衡细胞生长和生产。光遗传学提供了一种非侵入性,高度可调和可逆的动态调节基因表达的方法。在这里,我们描述了如何建立工程 大肠杆菌 和 酿酒酵母 的光控发酵,以生产化学物质或重组蛋白。我们讨论了如何在选定的时间和剂量下应用光,以分离微生物生长和生产,以改善发酵控制和生产率,以及获得最佳结果的关键优化考虑因素。此外,我们还描述了如何为实验室规模的生物反应器实验实施光控制。这些方案有助于在工程微生物中采用光遗传学控制,以提高发酵性能。
Introduction
光遗传学是利用光响应蛋白控制生物过程的一种新策略,为动态控制微生物发酵以进行化学和蛋白质生产提供了一种新策略1,2。工程代谢途径的负担以及某些中间体和产品的毒性通常会损害细胞生长3。这种压力会导致生物质积累不良并降低生产力3。这一挑战可以通过将发酵暂时划分为生长和生产阶段来解决,这些阶段将代谢资源分别用于生物质积累或产品合成4。我们最近表明,在这种两相发酵中,从生长到生产的过渡可以通过照明条件的变化来诱导5,6,7。光输入的高可调性、可逆性和正交性8 为光控发酵提供了独特的优势,而这些发酵剂很难或不可能用用于传统两相发酵动态控制的化学诱导剂复制4,9,10,11。
来自乳酸红杆菌的蓝光响应性EL222蛋白已被用于开发几种光遗传学电路,用于酿酒酵母的代谢工程5,7,12,13。EL222包含一个光氧电压传感器(LOV)结构域,该结构域在蓝光激活(465nm)时经历构象位移,使其能够与其同源DNA序列(C120)13结合。将EL222融合到病毒VP16激活结构域(VP16-EL222)可产生蓝光响应转录因子,该转录因子可逆地激活酿酒酵母7和其他生物体14中合成启动子PC120中的基因表达。基于EL222的几个电路已经开发并用于酿酒酵母的化学生产,例如基本的光激活OptoEXP系统7,其中目的基因直接从PC120表达(图1A)。然而,在发酵生产阶段通常遇到的高细胞密度下的光穿透问题促使我们开发在黑暗中诱导的倒置电路,例如OptoINVRT和OptoQ-INVRT电路(图1B)5,7,13。这些系统分别利用酿酒酵母和克拉沙猪笼草的半乳糖(GAL)或奎宁(Q)调节剂,用VP16-EL222控制其相应的抑制因子(GAL80和QS),以抑制光照下的基因表达,并在黑暗中强烈诱导。结合OptoEXP和OptoINVRT电路,可以双向控制基因表达,实现两相发酵,其中生长阶段由蓝光诱导,生产阶段与黑暗(图2A)5,7。
在生产阶段使用光而不是黑暗来诱导基因表达将大大扩展光遗传学控制的能力,但也需要克服在这个发酵阶段通常遇到的高细胞密度的光穿透限制。为此,我们开发了称为OptoAMP和OptoQ-AMP的电路,可以放大对蓝光刺激的转录响应。这些电路分别使用VP16-EL222的野生型或超敏突变体来控制GAL或Q调节子的转录激活剂Gal4p或QF2的产生,从而在光照下实现增强的灵敏度和更强的基因表达12,13(图1C)。OptoAMP电路可以在光密度(在600nm下测量;OD600)值至少为40,仅约0.35%的照度(仅在约7%的体积表面上为5%的光剂量)。与OptoEXP相比,这显示出更高的灵敏度,OptoEXP需要接近100%的照明12。在高细胞密度下用光有效诱导基因表达的能力为发酵的动态控制开辟了新的机会。这包括在两个以上的时间阶段进行发酵,例如三相发酵,其中生长,诱导和生产阶段以独特的轻计划建立,以优化化学生产(图2B)12。
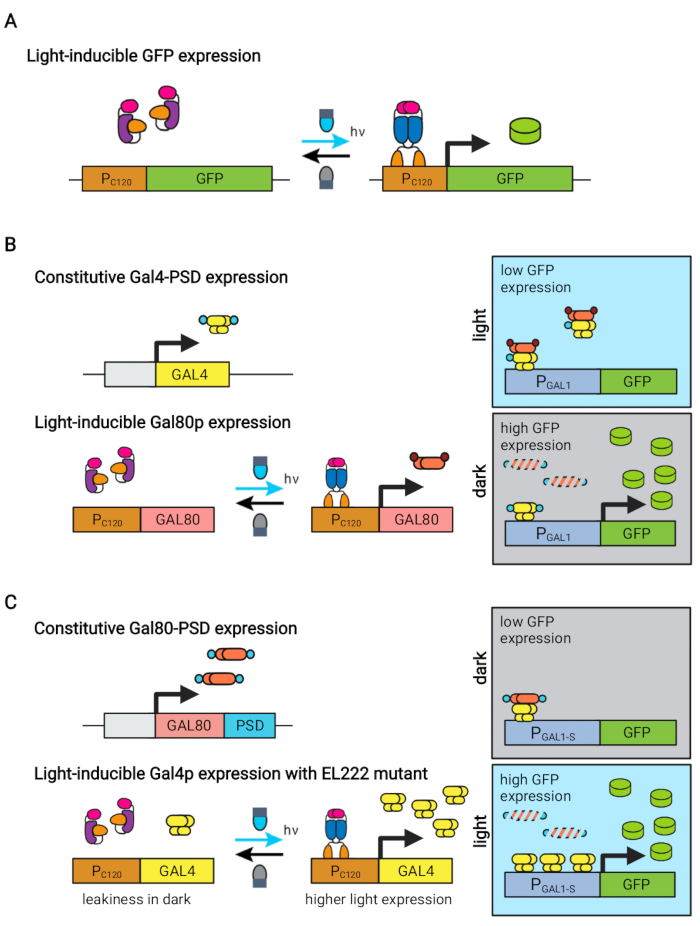
图1:用于动态控制酿酒酵母的光遗传学电路。 OptoEXP、OptoINVRT 和 OptoAMP 电路基于光敏 VP16-EL222 系统。(A)在OptoEXP电路中,暴露于蓝光会导致VP16-EL222的构象变化和二聚化,从而暴露DNA结合结构域并允许从PC120转录。该图由Zhao等人修改为7。(B) OptoINVRT电路利用GAL(如图所示)或Q调节子在黑暗中诱导表达。在基于GAL的电路中,VP16-EL222和GAL4是本构表达的,而PC120驱动GAL80抑制器的表达(在基于Q的电路中,GAL4和GAL80分别被QF2和QS取代,并且使用合成的含QUAS的启动子代替GAL启动子)。在光照下,Gal80p阻止了PGAL1目的基因的激活。在黑暗中,GAL80不被表达,并通过将其融合到一个本构的degron结构域(小的棕色结构域)而迅速降解,这允许Gal4p激活PGAL1。该图由Zhao等人修改为5。(C) 光电放大器电路还使用 VP16-EL222 来控制 GAL(如图所示)或 Q 重稳态。在这些电路中,GAL80抑制器(或QS)被组成表达并融合到光敏的degron(小蓝色域),确保在黑暗中进行严格的抑制。PC120和超敏的VP16-EL222突变体控制表达的GAL4(或QF2)与光,其强烈激活光中的PGAL1(或含QUAS的启动子)。GAL衍生的电路可以使用工程形式的PGAL1,例如PGAL1-M或PGAL1-S,它们具有增加的活性,以及由GAL调节子(PGAL1,PGAL10,PGAL2,PGAL7)控制的野生型启动子。该图由 Zhao 等人修改而来。请点击此处查看此图的放大版本。
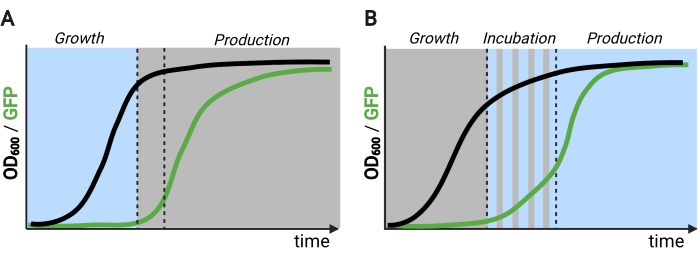
图 2:随时间推移的两相和三相发酵。 (A) 采用倒置回路操作的两相发酵包括光驱动的生长阶段和黑暗的生产阶段。在生长阶段,随着生产途径的抑制,生物质会积累。在达到所需的OD 600时,细胞被转移到黑暗中以代谢调整,然后重悬于新鲜培养基中以进行生产阶段。(B)在三阶段过程中,生长,孵化和生产阶段由独特的光照时间表定义,其中可能包括黑暗的生长期,脉冲孵育和完全照明的生产阶段。使用Biorender创建的图。 请点击此处查看此图的放大版本。
还开发了光遗传学电路,用于动态控制大肠杆菌中的化学和蛋白质产生。OptoLAC 电路使用基于 YF1/FixJ 双组分系统6 的光响应 pDawn 电路控制细菌 LacI 抑制器(图 3)。与OptoINVRT5类似,OptoLAC电路旨在抑制光中的基因表达并在黑暗中诱导基因表达。使用 OptoLAC 电路的表达水平可以达到或超过标准异丙基β-d-1-硫代乳糖焦氰胺苷 (IPTG) 诱导达到的水平,从而保持化学诱导的强度,同时提供增强的可调性和可逆性6。因此,OptoLAC电路能够为大肠杆菌的代谢工程提供有效的光遗传学控制。
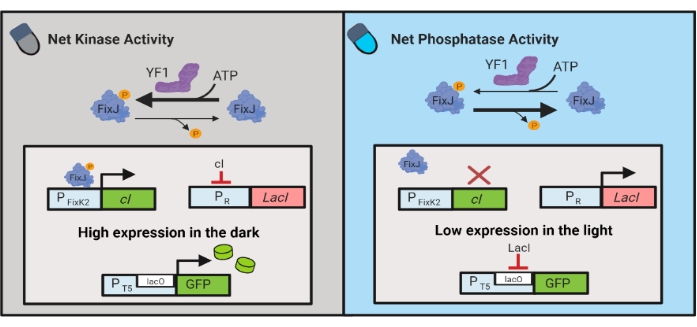
图 3:用于动态控制 大肠杆菌的 OptoLAC 电路。 OptoLAC电路调整pDawn系统和lac操纵子,以实现黑暗中的激活和光照下的抑制。在黑暗中,YF1磷酸化FixJ,然后激活PFixK2 启动子以表达 cI 抑制剂。 cI 抑制因子阻止来自PR启动子的 lacI 抑制因子的表达,PR 启动子允许从含 lacO的启动子转录目的基因。相反,蓝光降低YF1网激酶活性,逆转FixJ磷酸化,从而逆转 cI 表达,从而抑制 lacI 的表达并阻止含 lacO的启动子的表达。该图已从Lalwani等人6修改而来。 请点击此处查看此图的放大版本。
我们在这里描述了 酿酒酵母 和 大肠 杆菌用于化学或蛋白质生产的光控发酵的基本方案。对于酵母和细菌,我们首先关注由 OptoINVRT 和 OptoLAC 电路实现的光驱动生长阶段和黑暗诱导生产阶段的发酵。随后,我们描述了一种由OptoAMP电路实现的三相(生长,诱导,生产)光控发酵的协议。此外,我们还描述了如何将光遗传学控制的发酵从微孔板扩大到实验室规模的生物反应器。通过该协议,我们的目标是为化学或蛋白质生产进行光控发酵提供完整且易于重复的指南。
Protocol
Representative Results
Discussion
长期以来,动态控制一直被应用于提高代谢工程和重组蛋白生产的产量4。酶表达的转变通常使用化学诱导剂(如IPTG21,半乳糖22和四环素23)来实现,但也使用温度和pH等工艺条件介导。基因表达的光遗传学控制消除了改变发酵参数或培养基组成的需要,使其成为传统诱导策略的易于应用的替代方案。打开或关闭灯光的容易程…
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
这项研究得到了美国能源部,科学办公室,生物和环境研究办公室奖编号DE-SC0019363,NSF CAREER奖CBET-1751840,皮尤慈善信托基金和Camille Dreyfus教师学者奖的支持。
Materials
| Light-controlled chemical production using S. cerevisiae | |||
| 24-well culture plate | USA Scientific | CC7672-7524 | |
| Agar powder | Thermo Fisher Scientific | 303991049 | |
| Aluminum foil | Reynolds | B004NG90YO | |
| BioSpectrometer with μcuvette | Eppendorf | 6135000923 | |
| Blue LED panel | HQRP | 884667106091218 | |
| EZ-L439 OptoINVRT7 Plasmid | N/A | N/A | See Reference 1 |
| Glucose | Thermo Fisher Scientific | 501879892 (G8270-5KG) | |
| Microcentrifuge | Thermo Fisher Scientific | 75002403 | |
| Microcentrifuge tubes | USA Scientific | 1615-5510 | |
| Orbital Shaker | Yamato Scientific America | SOU-300 | |
| Petri dish | Celltreat | 229656 | |
| PmeI | New England Biolabs | R0560L | |
| Quantum meter | Apogee Instruments | MQ-510 | |
| Replica-plating device | Thomas Scientific | F37848-0000 | |
| Replica-plating pads | Sunrise Science Products | 3005-012 | |
| SC-His powder | Sunrise Science Products | 1303-030 | |
| SC Complete powder | Sunrise Science Products | 1459-100 | |
| Sterile sealing film | Excel Scientific | STR-SEAL-PLT | |
| YPD agar plates | VWR | 100217-054 | |
| Zeocin | Thermo Fisher Scientific | R25005 | |
| Light-controlled protein production using E. coli | |||
| 6X SDS Sample Buffer | Cepham Life Sciences | 10502 | |
| 12% Acrylamide protein gels | Thermo Fisher Scientific | NP0341BOX | |
| 24-well culture plate | USA Scientific | CC7672-7524 | |
| Aluminum foil | Reynolds | B004NG90YO | |
| BioSpectrometer with μcuvette | Eppendorf | 6135000923 | |
| Blue LED panel | HQRP | 884667106091218 | |
| Coomassie Brilliant Blue G-250 | Thermo Fisher Scientific | 20279 | |
| Electrophoresis cell | Bio-Rad | 1658004 | |
| Electrophoresis power supply | Bio-Rad | 1645050 | |
| LB broth (Miller) | Fisher Scientific | BP97235 | |
| Microcentrifuge | Thermo Fisher Scientific | 75002403 | |
| Microcentrifuge tubes | USA Scientific | 1615-5510 | |
| NaCl | Thomas Scientific | SX0425-1 | |
| OptoLAC plasmids | N/A | N/A | See Reference 2 |
| Orbital Shaker | Yamato Scientific America | SOU-300 | |
| Petri dish | Celltreat | 229656 | |
| Quantum meter | Apogee Instruments | MQ-510 | |
| SOC medium | Thermo Fisher Scientific | 15544034 | |
| Thermomixer | Eppendorf | 5382000015 | |
| Tris base | Fisher Scientific | BP1521 | |
| Three-phase fermentation using S. cerevisiae | |||
| Same materials as "Light-controlled chemical production using S. cerevisiae" protocol plus the following: | |||
| EZ-L580 OptoAMP4 Plasmid | N/A | N/A | See Reference 10 |
| Chemical production in a light-controlled bioreactor | |||
| Aluminum foil | Reynolds | B004NG90YO | |
| Antifoam | Sigma-Aldrich | A8311 | |
| Bioreactor with control station | Eppendorf | B120110001 | |
| BioSpectrometer with μcuvette | Eppendorf | 6135000923 | |
| Bleach | VWR Scientific | 89501-620 (CS) | |
| Blue LED panel | HQRP | 884667106091218 | |
| BPT tubing | Fisher Scientific | 14-170-15 | |
| Glucose | Thermo Fisher Scientific | 501879892 (G8270-5KG) | |
| Hydrochloric acid (HCl) | Fisher Scientific | 7647-01-0 | |
| M9 Minimal Salts | Thermo Fisher Scientific | A1374401 | |
| Microcentrifuge | Thermo Fisher Scientific | 75002403 | |
| Microcentrifuge tubes | USA Scientific | 1615-5510 | |
| NH4OH Solution | Sigma-Aldrich | I0503-1VL | |
| Orbital Shaker | Yamato Scientific America | SOU-300 | |
| Quantum meter | Apogee Instruments | MQ-510 | |
| SC Complete powder | Sunrise Science Products | 1459-100 |
Riferimenti
- Figueroa, D., Rojas, V., Romero, A., Larrondo, L. F., Salinas, F. The rise and shine of yeast optogenetics. Yeast. 38 (2), 131-146 (2021).
- Pouzet, S., et al. The promise of optogenetics for bioproduction: Dynamic control strategies and scale-up instruments. Bioingegneria. 7 (4), 151 (2020).
- Venayak, N., Anesiadis, N., Cluett, W. R., Mahadevan, R. Engineering metabolism through dynamic control. Current Opinion in Biotechnology. 34, 142-152 (2015).
- Lalwani, M. A., Zhao, E. M., Avalos, J. L. Current and future modalities of dynamic control in metabolic engineering. Current Opinion in Biotechnology. 52, 56-65 (2018).
- Zhao, E. M., et al. Design and characterization of rapid optogenetic circuits for dynamic control in yeast metabolic engineering. ACS Synthetic Biology. 9 (12), 3254-3266 (2020).
- Lalwani, M. A., et al. Optogenetic control of the lac operon for bacterial chemical and protein production. Nature Chemical Biology. 17 (1), 71-79 (2021).
- Zhao, E. M., et al. Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature. 555 (7698), 683-687 (2018).
- Baumschlager, A., Khammash, M. Synthetic biological approaches for optogenetics and tools for transcriptional light-control in bacteria. Advanced Biology. 5 (5), 2000256 (2021).
- Dvorak, P., et al. Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21(DE3) carrying a synthetic metabolic pathway. Microbial Cell Factories. 14, 201 (2015).
- Hartline, C. J., Schmitz, A. C., Han, Y., Zhang, F. Dynamic control in metabolic engineering: Theories, tools, and applications. Metabolic Engineering. 63, 126-140 (2021).
- Ni, C., Dinh, C. V., Prather, K. L. J. Dynamic control of metabolism. Annual Review of Chemical and Biomolecular Engineering. 12, 519-560 (2021).
- Zhao, E. M., et al. Optogenetic amplification circuits for light-induced metabolic control. ACS Synthetic Biology. 10 (5), 1143-1154 (2021).
- Lalwani, M. A., Zhao, E. M., Wegner, S. A., Avalos, J. L. The Neurospora crassa Inducible Q System Enables Simultaneous Optogenetic Amplification and Inversion in Saccharomyces cerevisiae for Bidirectional Control of Gene Expression. ACS Synthetic Biology. 10 (8), 2060-2075 (2021).
- Motta-Mena, L. B., et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nature Chemical Biology. 10 (3), 196-202 (2014).
- Gietz, R. D., Woods, R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods in Enzymology. 350, 87-96 (2002).
- Marx, H., Mecklenbräuker, A., Gasser, B., Sauer, M., Mattanovich, D. Directed gene copy number amplification in Pichia pastoris by vector integration into the ribosomal DNA locus. FEMS Yeast Research. 9 (8), 1260-1270 (2009).
- Nordén, K., et al. Increasing gene dosage greatly enhances recombinant expression of aquaporins in Pichia pastoris. BMC Biotechnology. 11, 47 (2011).
- Zhao, E. M., et al. Light-based control of metabolic flux through assembly of synthetic organelles. Nature Chemical Biology. 15 (6), 589-597 (2019).
- Dowee, W. J., Miller, J. F., Ragsdale, C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Research. 16 (13), 6127-6145 (1988).
- Zhou, K., Edgar, S., Stephanopoulos, G. Engineering microbes to synthesize plant isoprenoids. Methods in Enzymology. 575, 225-245 (2016).
- Arfman, N., Worrell, V., Ingram, L. O. Use of the tac promoter and lacI(q) for the controlled expression of Zymomonas mobilis fermentative genes in Escherichia coli and Zymomonas mobilis. Journal of Bacteriology. 174 (22), 7370-7378 (1992).
- Steen, E. J., et al. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microbial Cell Factories. 7 (1), 1-8 (2008).
- Tan, S. Z., Manchester, S., Prather, K. L. J. Controlling central carbon metabolism for improved pathway yields in Saccharomyces cerevisiae. ACS Synthetic Biology. 5 (2), 116-124 (2015).
- Jayaraman, P., et al. Blue light-mediated transcriptional activation and repression of gene expression in bacteria. Nucleic Acids Research. 44 (14), 6994 (2016).
- Fernandez-Rodriguez, J., Moser, F., Song, M., Voigt, C. A. Engineering RGB color vision into Escherichia coli. Nature Chemical Biology. 13 (7), 706-708 (2017).
- Ding, Q., et al. Light-powered Escherichia coli cell division for chemical production. Nature Communications. 11 (1), 1-14 (2020).
- Senoo, S., Tandar, S. T., Kitamura, S., Toya, Y., Shimizu, H. Light-inducible flux control of triosephosphate isomerase on glycolysis in Escherichia coli. Biotechnology and Bioengineering. 116 (12), 3292-3300 (2019).
- Ramakrishnan, P., Tabor, J. J. Repurposing synechocystis PCC6803 UirS-UirR as a UV-violet/green photoreversible transcriptional regulatory tool in E. Coli. ACS Synthetic Biology. 5 (7), 733-740 (2016).
- Tabor, J. J., Levskaya, A., Voigt, C. A. Multichromatic control of gene expression in escherichia coli. Journal of Molecular Biology. 405 (2), 315-324 (2011).
- Stewart, C. J., McClean, M. N. Design and implementation of an automated illuminating, culturing, and sampling system for microbial optogenetic applications. Journal of Visualized Experiments:JoVE. (120), e54894 (2017).
- Grødem, E. O. S., Sweeney, K., McClean, M. N. Automated calibration of optoPlate LEDs to reduce light dose variation in optogenetic experiments. BioTechniques. 69 (4), 313-316 (2020).
- Gerhardt, K. P., et al. An open-hardware platform for optogenetics and photobiology. Scientific Reports. 6, (2016).
- Bugaj, L. J., Lim, W. A. High-throughput multicolor optogenetics in microwell plates. Nature Protocols. 14 (7), 2205-2228 (2019).
- Steel, H., Habgood, R., Kelly, C., Papachristodoulou, A. In situ characterisation and manipulation of biological systems with Chi.Bio. PLoS Biology. 18 (7), (2020).
- Carrasco-López, C., García-Echauri, S. A., Kichuk, T., Avalos, J. L. Optogenetics and biosensors set the stage for metabolic cybergenetics. Current Opinion in Biotechnology. 65, 296-309 (2020).
- Milias-Argeitis, A., Rullan, M., Aoki, S. K., Buchmann, P., Khammash, M. Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth. Nature Communications. 7 (1), 1-11 (2016).
- Melendez, J., et al. Real-time optogenetic control of intracellular protein concentration in microbial cell cultures. Integrative Biology: Quantitative Biosciences From Nano to Macro. 6 (3), 366-372 (2014).
- Milias-Argeitis, A., et al. In silico feedback for in vivo regulation of a gene expression circuit. Nature Biotechnology. 29 (12), 1114-1116 (2011).
- Castillo-Hair, S. M., Baerman, E. A., Fujita, M., Igoshin, O. A., Tabor, J. J. Optogenetic control of Bacillus subtilis gene expression. Nature Communications. 10 (1), 1-11 (2019).
- Xia, A., et al. Optogenetic modification of pseudomonas aeruginosa enables controllable twitching motility and host infection. ACS Synthetic Biology. 10 (3), 531-541 (2021).
- Pu, L., Yang, S., Xia, A., Jin, F. Optogenetics manipulation enables prevention of biofilm formation of engineered pseudomonas aeruginosa on surfaces. ACS Synthetic Biology. 7 (1), 200-208 (2018).

