A Novel High-Throughput Ex Vivo Ovine Skin Wound Model for Testing Emerging Antibiotics
Summary
The protocol describes a step-by-step method to set up an ex vivo ovine wounded skin model infected with Staphylococcus aureus. This high-throughput model better simulates infections in vivo compared with conventional microbiology techniques and presents researchers with a physiologically relevant platform to test the efficacy of emerging antimicrobials.
Abstract
The development of antimicrobials is an expensive process with increasingly low success rates, which makes further investment in antimicrobial discovery research less attractive. Antimicrobial drug discovery and subsequent commercialization can be made more lucrative if a fail-fast-and-fail-cheap approach can be implemented within the lead optimization stages where researchers have greater control over drug design and formulation. In this article, the setup of an ex vivo ovine wounded skin model infected with Staphylococcus aureus is described, which is simple, cost-effective, high throughput, and reproducible. The bacterial physiology in the model mimics that during infection as bacterial proliferation is dependent on the pathogen's ability to damage the tissue. The establishment of wound infection is verified by an increase in viable bacterial counts compared to the inoculum. This model can be used as a platform to test the efficacy of emerging antimicrobials in the lead optimization stage. It can be contended that the availability of this model will provide researchers developing antimicrobials with a fail-fast-and-fail-cheap model, which will help increase success rates in subsequent animal trials. The model will also facilitate the reduction and refinement of animal use for research and ultimately enable faster and more cost-effective translation of novel antimicrobials for skin and soft tissue infections to the clinic.
Introduction
Skin infections are an important global issue, with large economic costs to healthcare providers around the world. The development of multidrug resistance and biofilm formation by pathogens plays a key role in the prevalence of non-healing wounds1,2,3,4. As a result of this, skin and soft tissue infections are one of the more common reasons for extended hospitalization and subsequent readmission5. Delays in wound healing are costly for both the patient and healthcare providers, with some estimates suggesting around 6.5 million patients are affected annually in the US. In the UK, skin infections and associated complications result in approximately 75,000 deaths annually2,4,6.
Staphylococcus aureus (S. aureus) is a formidable wound pathogen frequently isolated from patient wounds2,7. The rate of emergence of multidrug resistance increased drastically in the 2000s. During this time, around 60% of acute bacterial skin and skin structure infections were culture positive for methicillin-resistant S. aureus1. The increasing number of multidrug-resistant strains among Staphylococci, and indeed other pathogens, within the last 2 decades indicates an urgent need for the rapid development of antibiotics with new modes of action that can overcome resistance.
However, since the early 2000s, antibiotic discovery programs have been dominated by longer developmental times and low success rates, with only 17% of novel antibiotics entering clinical trials in the US achieving market approval8. This suggests a disparity between results from in vitro testing of emerging antibiotics and their clinical outcomes. It can be contended that this disparity is largely due to differences in bacterial physiology during infections in vivo and during conventional microbiological methods when testing the efficacy of antibiotics in the in vitro preclinical stages. Therefore, novel laboratory methods that are more representative of bacterial physiology during infection are needed to improve the success rates in antibiotic discovery programs.
Current methods for studying skin infections include studies in live animals (e.g., mice), ex vivo skin models (e.g., porcine), and 3D tissue-engineered skin models (e.g., human)9,10,11,12. Studies in live animals are strictly regulated and have relatively low throughput. In animal models, wounding and infection cause significant distress to the animals and raise ethical concerns. Human skin models, ex vivo or tissue-engineered, require ethical approval, compliance with local and global legislation (the Human Tissue Act, the Declaration of Helsinki), and there is difficulty in acquiring tissues, with some requests taking years to fulfil13,14. Both model types are labor intensive and require significant expertise to ensure experimental success. Some current ex vivo skin infection models require pre-inoculated discs and additives for the wound bed to enable infection; although these models are incredibly useful, there are limitations in the infection process as additives limit the utilization of the wound bed as a nutrient source10,15,16,17. The model described in this study uses no additives to the wound bed, which ensures that the pathology of infection and viable cell counts are a result of direct utilization of the wound bed as the only nutrient source.
Given the need for new laboratory methods, a novel high-throughput ex vivo ovine model of skin infections for use in evaluating the efficacy of emerging antibiotics has been developed. Skin infection studies face many challenges-high costs, ethical concerns, and models that do not show a full picture20,21. Ex vivo models and 3D explant models allow for better visualization of the disease process and the impact treatments can have from a more clinically relevant model. Here, the setup of a novel ovine skin model is described, which is simple, reproducible, and clinically relevant and has high throughput. Ovine skin was chosen as sheep are one of the large mammals commonly used to model responses to infections in vivo. Moreover, they are readily available from abattoirs, ensuring a steady supply of skin for research, and their carcasses are not scalded, ensuring good tissue quality. This study used S. aureus as the exemplar pathogen; however, the model works well with other microorganisms.
Protocol
Lambs' heads from the R.B Elliott and Son Abattoir were used as the source of skin samples in this project. All lambs were slaughtered for consumption as food. Instead of discarding the heads, these were repurposed for research. Ethics approval was not required as the tissue was sourced from waste discarded from abattoirs.
1. Sterilization
- Disinfect forceps prior to collection of the heads by taking clean forceps and performing dry heat sterilization in an oven at 200 °C for 1 h. Autoclave all glassware at 121 °C for 15 min before use.
- Carry out all described work within a microbiology class 2 cabinet. Prepare all reagents as per the manufacturer's instructions.
2. Sample collection
- In the abattoir, Swaledale lambs were slaughtered by stunning using electricity or a captive-bolt pistol and exsanguinated. Collect lamb heads no more than 4 h post-slaughter.
3. Preparation of the heads
- Disinfect the forehead section of the lamb by pouring approximately 100 mL of 200 ppm chlorine dioxide solution onto the sample area. Shave the forehead section of the head using electric clippers and wash the area with 200 mL of 200 ppm chlorine dioxide solution.
- Wipe the area with ethanol and blue roll and cover the sample area with hair removal cream for 35 min. Gently scrape off the hair removal cream using a scraping tool and assess the sample area. If a significant amount of hair remains, repeat the hair removal process.
- Use a further 200 mL of chlorine dioxide solution to rinse the area, and then rinse with ethanol and wipe with a blue roll.
- Using a sterile 8 mm biopsy punch, cut out 8 mm skin samples from the prepared area. Remove the samples using sterile forceps and a 15-blade scalpel, ensuring that all cutaneous fat is removed.
- Place the samples in a sterile 0.5 L jar filled with sterile phosphate-buffered saline (PBS), and then transfer them to sterile 50 mL tubes with 50 mL of 200 ppm chlorine dioxide solution, invert twice, and leave to sterilize for 30 min.
- Remove the samples from the chlorine dioxide solution and wash them by placing them in a 50 mL tube filled with 40 mL of sterile PBS. Once washed, place each individual skin sample in a separate well of a 24-well plate.
- Add 350 µL of pre-warmed medium while maintaining the sample at the air-liquid interface. The composition of the media is as follows: MK media (Medium 199 with Hanks' salts, L-glutamate, and 1.75 mg/mL sodium bicarbonate) and Ham's F12 in a 1:1 ratio, with added FBS (10% v/v), EGF (10 ng/mL), insulin (5 µg/mL), penicillin-streptomycin (100 U/mL), and amphotericin B (2.5 µg/mL).
- Seal the 24-well plates with a gas permeable plate seal and incubate at 37 °C in a humidified 5% CO2 tissue incubator for up to 24 h.
4. Maintenance of skin samples
- After incubation, remove the culture medium and rinse the samples in 500 µL of sterile PBS. Add antibiotic-free media to each sample and incubate at 37 °C in a humidified 5% CO2 tissue incubator for 24 h to remove residual antibiotics in the sample.
- If turbidity or fungal infection develop in the antibiotic-free media after 24 h, then discard the sample.
5. Preparation of the inoculum
- Prepare a 50 mL tube with 10 mL of sterile tryptic soy broth. Take a fresh agar plate of S. aureus and use a swab to transfer several colonies into the broth. Incubate for 18 h at 37 °C at 150 rpm.
- Centrifuge at 4,000 x g for 3 min. Remove the supernatant and resuspend the cell pellet in 10 mL of sterile PBS. Repeat twice to ensure adequate washing of the cells.
- Adjust the inoculum to 0.6 OD600 in sterile PBS. Confirm the inoculum load by undertaking a manual viable plate count.
6. Infection of skin samples
- Prepare a fresh 24-well plate with 400 µL of pre-warmed antibiotic-free media and add in the 24-well inserts using sterile forceps.
- Remove the media from the skin samples, wash with 500 µL of sterile PBS, and remove the wash. Use sterile forceps to gently hold the sample to the bottom of the well.
- Use a 4 mm punch biopsy to make a central wound flap, piercing through to a rough depth of 1-2 mm. Then, use a 15-blade scalpel and sterile toothed allis tissue forceps to remove the top layer of the wound flap. Variability in the wound dimensions may affect the outcome of the infection and the endpoint colony forming units (CFU).
- Once all the samples have been wounded, transfer them to the 24-well inserts using sterile forceps. Pipette 15 µL of the bacterial inoculum into the wound bed. Then, incubate for 24 h at 37 °C in a humidified 5% CO2 tissue incubator.
- If longer incubation periods are necessary, remove the media and replace it with fresh media every 24 h and incubate in the same conditions.
7. Determination of the bacterial load
- Remove the media from the bottom of the wells. With sterile forceps, transfer each sample into a separate 50 mL tube filled with 1 mL of sterile PBS.
- Use a fine-tipped homogenizer to homogenize the surface of the sample. Take care to ensure that the wound bed is in direct contact with the tip of the homogenizer.
- Homogenize each sample for 35 s on medium/high. The homogenizer detaches the bacteria from the surface of the wound bed to allow for enumeration of the bacterial load.
- Once all the samples are processed, vortex each sample, in turn, prior to pipetting. This is to ensure the bacterial homogenate is mixed.
- Pipette 20 µL of vortexed homogenate into the corresponding well of a 96-well plate containing 180 µL of sterile PBS.
- Serially dilute each sample homogenate to 1 x 10−7 and pipette 10 µL of the diluted homogenate onto a tryptic soy agar plate in triplicate.
- Incubate the agar plate for 18 h at 37 °C. Then, count the number of colonies to determine the CFU for each sample.
Representative Results
The identification of a route to sterilize the skin before setting up the wound infection model was challenging. The challenge lay in sterilizing the skin without damaging the different skin layers, which may then go on to have unintended consequences in the outcome of infection. To identify an appropriate sterilization regime, different treatments were tried for varying lengths of time, as outlined in Table 1. Contamination was recorded as the development of turbidity after 48 h in the MK medium used to maintain the skin samples. Tissue integrity was monitored by histology followed by staining with hematoxylin and eosin (H&E) immediately after treatment (Figure 1). A 30 min treatment with chlorine dioxide proved the most effective at reproducibly sterilizing the skin tissue whilst preserving tissue integrity.
In the experimental setup, the wounded sterile tissue is placed in the apical chamber of a 24-well insert at the air-liquid interface (Figure 2A). Two different wounding techniques were attempted-a flap removal technique, in which the tissue is wounded by a punch biopsy tool and the top layer of the wounded tissue is removed using a combination of a 15-blade scalpel and sterile toothed allis tissue forceps, and a scratch technique, in which the tissue is wounded by a punch biopsy tool alone. Although the scratch model harbored a higher average number of CFUs after 24 h, the results were variable. The flap removal technique produced more consistent results (Figure 2C). After 48 h, approximately 100 times more bacterial CFUs were recovered from the wounded tissue compared to the inoculum (Figure 2D). This was deemed to indicate a successful infection. A white film in the wound bed was evident on the tissue 48 h after infection (Figure 2B). Gram-stained histology sections of infected tissue indicated the presence of S. aureus cells in the wound bed (Figure 2E,F). Gram-stained histology sections of uninfected tissue are provided as a comparator (Figure 2G,H).
From one lamb's head, a 100 cm2 section (10 cm x 10 cm) can be realistically accessed. Given each patch of skin is punched out of the forehead using a circular 8 mm diameter punch biopsy, approximately 100 skin patches can be obtained from one lamb's forehead per week. Procuring further lambs' heads proportionately increases the number of skin samples that can be obtained per week. Hence, it is claimed that the procedure is relatively high throughput. For this study, 24 skin samples were obtained routinely per week. Skin from various lambs' heads was processed as biological replicates to account for the potential donor to donor variability. During the preparation of the skin patches (steps 3.1-3.8), the major time consumption is the 30 min incubation of the skin samples in the chlorine dioxide solution and the 2 x 35 min wait for the hair removal cream treatment. The use of punch biopsy to generate the 24 skin patches takes about 15 min. It is this time that would scale linearly if the number of skin patches were increased.
| Disinfectant | 10 Min | 30 Min | 60 Min |
| 1% high-level medical surface disinfectant | F | P | P |
| 1% multi-purpose disinfectant | F | F | P |
| 3% povidone | F | F | F |
| 5% povidone | F | P | P |
| 10% povidone | F | P | P |
| 70% ethanol | F | P | P |
| UV | F | F | F |
| 0.55% hypochlorite | F | P | P |
| 200 ppm chlorine dioxide | F | P | P |
Table 1: Sterilization of fresh lamb skin. Each skin sample was left in the disinfectant for the specified time. F denotes failure to sterilize the tissue, with bacterial contamination present in the media. P denotes passing, with no bacterial contamination present in the media.
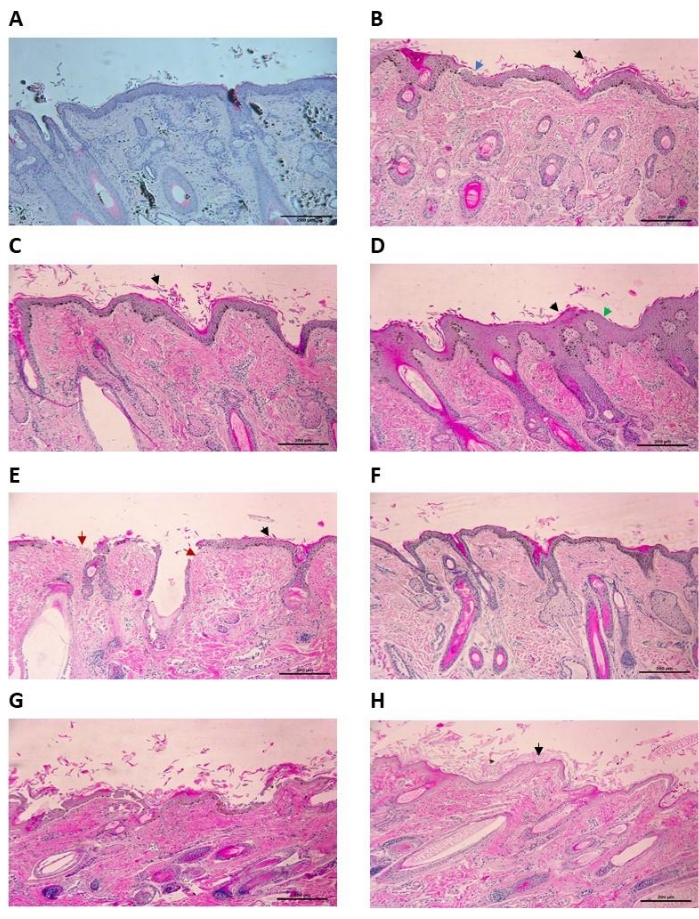
Figure 1: Histology of uninfected ex vivo ovine skin treated with disinfectants (H&E stain) at 100x magnification. (A) Control skin sample with 30 min treatment in PBS. Some epidermal shedding can be seen, but the epidermis is not disrupted. (B) Ovine skin treated with 1% high-level medical surface disinfectant for 30 min. Epidermal shedding (black arrow) along with some disruption to the stratum granulosum (blue arrow). (C) Ovine skin treated with 5% povidone for 30 min. Moderate damage to the tissue can be seen here, with epidermal shedding of the stratum corneum (black arrow). There is minimal disruption to the underlying epidermal layers. (D) Ovine skin treated with 10% povidone for 30 min. The top layers of the epidermis have been damaged, with evidence of shedding (black arrow) and thinning (green arrow). (E) Ovine skin treated with 2% multi-purpose disinfectant for 30 min. Severe damage to the sample can be observed, with significant epidermal shedding (black arrow) and complete eradication of the stratum corneum (red arrow). (F) Ovine skin treated in 200 ppm chlorine dioxide for 30 min. Some epidermal shedding can be seen, but the epidermis is intact. (G) Ovine skin treated with 0.6% hypochlorite for 30 min. Severe damage to the epidermis is present, with a high level of epidermal shedding (black arrow) and eradication of the epidermis in places (red arrow). (H) Ovine skin treated with 70% ethanol for 30 min. Damage to the epidermis can be seen, with significant epidermal shedding (black arrow). Scale bar is 200 µm. Please click here to view a larger version of this figure.
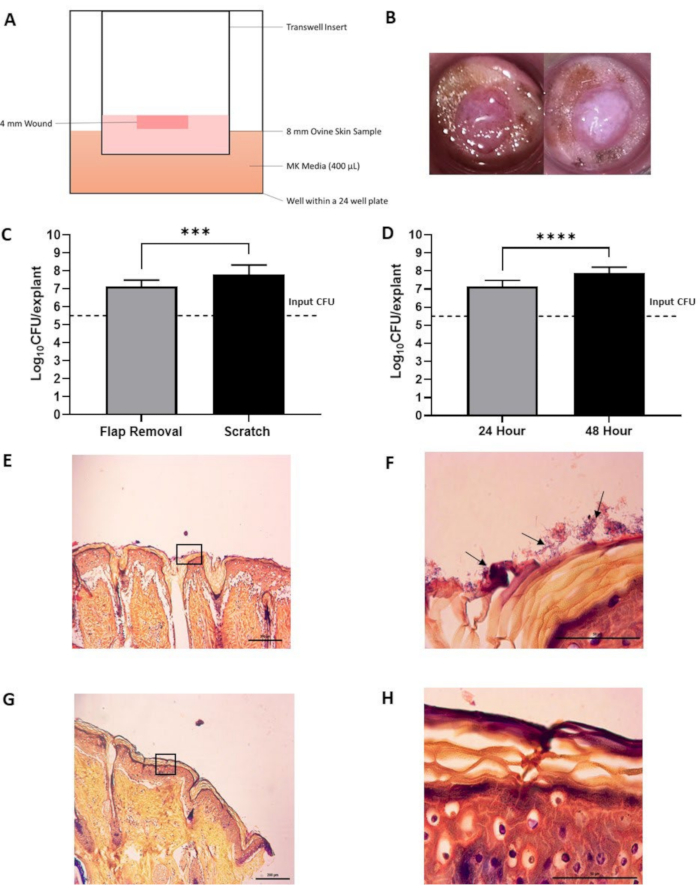
Figure 2: Ex vivo ovine skin infected with S. aureus. (A) Schematic of the experimental setup. (B) Pictures of ex vivo ovine skin prior to infection (left image) and post 48 h infection (right image). Note the white film present following 48 h incubation in infected tissue, which is absent in uninfected tissue. (C) Testing the effect of different wounding methods on the final colony forming unit (CFU) counts after homogenization. Flap removal (n = 6) and scratch (n = 9) infected with S. aureus for 24 h. Error bars indicate standard deviation. *** indicates a p-value < 0.0001. (D) Testing the effect of incubation times on CFU counts following homogenization. 24 h incubation (n = 6) and 48 h incubation (n = 12). Error bars indicate standard deviation. **** indicates a p-value < 0.0001. (E,F) Representative images of histopathological analysis of infected ovine skin post 48 h infection with a modified Gram stain (E) at 100x magnification (scale bar of 200 µm). The box indicates the area magnified in (F) at 1,000x magnification (scale bar of 50 µm). Black arrows indicate bacteria. (G,H) Representative images of histopathological analysis of uninfected ovine skin following a 48 h period (control) with a modified gGram stain (G) at 100x magnification (scale bar of 200 µm). The box indicates the area magnified in (H) at 1,000x magnification (scale bar of 50 µm). Statistical analysis was carried out as a one-way ANOVA. Please click here to view a larger version of this figure.
Discussion
The development of antimicrobials is an important but expensive venture that is estimated to cost around $1 billion and take around 15 years to complete. Over 90% of antimicrobial drug discovery and preclinical studies of antimicrobial drug efficacy are carried out by academic researchers and small to medium companies with typically less than 50 employees22. These teams are very financially constrained, which makes the failure of lead molecules in later stages of translational research calamitous. The rise in antimicrobial resistance is outpacing the development of novel antimicrobials, which further intensifies the need to steward the already limited investment in antimicrobial research responsibly23.
The disparity in bacterial physiology between infections in in vivo and in vitro laboratory cultures tends to cause an overestimation of the antimicrobial efficacy of the hits during the lead identification stage. Such overestimation contributes to the high attrition rates seen during the subsequent stages of translation. The availability of in vitro antimicrobial efficacy testing platforms that incorporate bacteria with a physiology that better mimics that found during infections in vivo will enable a more accurate estimation of antimicrobial efficacy and provide researchers with greater control of lead optimization without reliance on expensive and tightly regulated animal trials. Such models will also decrease attrition rates during the subsequent stages of translation and may make antimicrobial drug discovery more attractive for investment.
This ovine skin infection model described here is a useful tool that will provide researchers with a physiologically relevant in vitro model of an infected wound to test the efficacy of emerging topical antimicrobial formulations at the preclinical stages. In contrast to the majority of ex vivo infection models described in the scientific literature10,15,24,25,26,27,28, in this model, the pathogens exposed to the ex vivo tissue are not supplemented with culture media during infection. Pathogen proliferation and subsequent infection are dependent on the ability of the organism to damage the tissue. Therefore, the pathogen physiology in this model is more closely related to that during in vivo infections compared to conventional microbiological cultures. A highly reproducible infection is obtained from this model, as judged by the number of CFUs retrieved from the tissue after incubation.
With the plethora of human skin equivalents and human and porcine ex vivo wound models, the use of ovine tissue in this model could be superficially argued to be a limitation. However, sheep are among the group of large animals used as models of infections in vivo29,30,31. Moreover, sheep have been used as surrogates for humans in immunology research and vaccine development, indicating that their immune responses are similar to those of humans32. The tissue repair process during wound healing is similar in large mammals, including sheep and humans33,34, and interventions to improve wound healing that have been successfully demonstrated in sheep have been suggested for translation to humans35,36,37. Therefore, the use of ex vivo ovine skin in this wound model is not a limitation. Furthermore, this ovine model can be judged to be a reliable alternative to models incorporating human or porcine skin for the following reasons. The availability of human skin is limited and of variable quality when available38. Tissue-engineered human epidermis and living skin equivalents require refinements in their culturing conditions to ensure reproducibility in tissue physiology39. Although porcine skin is more accessible than human skin, the widely available porcine skin is scalded as part of carcass processing in the abattoir, which removes the epidermis10. From experience, unscalded porcine skin is less readily available from abattoirs.
Throughout the protocol described here, there are critical steps that ensure the success of the model. The most critical step involves the sterilization of the tissue following harvesting. Chlorine dioxide solution must be mixed with the tissue samples, and a contact time of 30 min must be allowed to sufficiently disinfect the tissue and remove contaminants. Furthermore, when setting up this experiment, turbidity or fungal contamination may occur due to improper handling or ineffective treatment with disinfectants and antibiotic media. In such a case, samples that develop turbidity must be discarded. To reduce the development of turbidity and contamination, the laboratory environment should be kept clean, with frequent sterilization of the incubator, the use of filter tips, and ensuring complete sterilization of the tools and containers used with the samples. Another critical step in the protocol is when the 4 mm biopsy punch is used to wound the tissue. The use of this tool allows for similarly sized wound beds to ensure the repeatability and reproducibility of the protocol. Variability in the wound bed size impacts the endpoint CFU. When there is adequate removal of the tissue flap, the results are much more consistent. A further critical step associated with the integrity of the endpoint CFU count is the use of the fine-tipped homogenizer to liberate bacterial cells. The position of the homogenizer tip was seen to have an impact on the CFU count from sample to sample; when the tip was correctly placed directly on the wound bed, there was much less variability seen from sample to sample.
Nevertheless, the model proposed here is not without limitations. This model has limitations inherent to all ex vivo studies (i.e., the lack of active vascular flow, the absence of commensal microbiota, which may modulate infection progress, and the absence of immune cells). It can be argued that, since the ex vivo wound model does not incorporate active immune cells, the infection progression in vivo in the presence of these cells could be different from that observed in ex vivo models. Regardless, ex vivo models provide a tissue surface for attachment and a nutrient source for bacteria and present a 3D diffusion barrier to formulations, which enables a more accurate assessment of antimicrobial efficacy than conventional microbiology culture techniques.
A key advantage of the model is that the tissue is sourced from lambs that are grown for human consumption. More particularly, the model repurposes skin from lambs' heads, which are usually discarded. Moreover, the model has high throughput and enables cost-effective, rapid, and reproducible comparative studies. It can be contended that the availability of this model will reduce and refine the need for animals purpose-bred for translational research in line with the principles of the three Rs, which facilitate more humane research practices. Although the model described here incorporates S. aureus as the exemplar pathogen, the model can be used with other microorganisms including other bacteria, fungi, and viruses, thus broadening the scope of drug development that the model enables. It can be envisioned that the use of this model will enable the rapid translation of much-needed antibiotics for skin infections by providing researchers with greater control over drug design and formulation at the preclinical stages.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank EPSRC (EP/R513313/1) for funding. The authors would like to also thank R.B Elliot and Son Abattoir in Calow, Chesterfield, for providing lambs' heads and for being so accommodating in the early stages of the project, Kasia Emery for her support throughout the development of this protocol, and Fiona Wright from the Department of Infection, Immunity and Cardiovascular Disease at the University of Sheffield for processing the histology samples and being so incredibly helpful throughout this project.
Materials
| 24 Well Companion Plate | SLS | 353504 | |
| 4 mm Biopsy Punch | Williams Medical | D7484 | |
| 50 ml centrifuge tubes | Fisher Scientific | 10788561 | |
| 8 mm Biopsy Punch | Williams Medical | D7488 | |
| Amphotericin B solution, sterile | Sigma | A2942 | |
| Colour Pro Style Cordless Hair Clipper | Wahl | 9639-2117X | Hair Clippers |
| Dual Oven Incubator | SLS | OVe1020 | Sterilising oven |
| Epidermal growth factor | SLS | E5036-200UG | |
| Ethanol | Honeywell | 458600-2.5L | |
| F12 HAM | Sigma | N4888 | |
| Foetal bovine serum | Labtech International | CA-115/500 | |
| Forceps | Fisher Scientific | 15307805 | |
| Hair Removal Cream | Veet | Not applicable | |
| Heracell VIOS 160i | Thermo Scientific | 15373212 | Tissue culture incubator |
| Heraeus Megafuge 16R | VWR | 521-2242 | Centrifuge |
| Homogenizer 220, Handheld | Fisher Scientific | 15575809 | |
| Homogenizer 220, plastic blending cones | Fisher Scientific | 15585819 | |
| Insert Individual 24 well 0.4um membrane | VWR International | 353095 | |
| Insulin, recombinant Human | SLS | 91077C-1G | |
| Medium 199 (MK media) | Sigma | M0393 | |
| Microplate, cell culture Costar 96 well | Fisher Scientific | 10687551 | |
| Multitron | Infors | Not applicable | Bacterial incubator |
| PBS tablets | Sigma | P4417-100TAB | |
| Penicillin-Streptomycin | SLS | P0781 | |
| Plate seals | Fisher Scientific | ESI-B-100 | |
| Safe 2020 | Fisher Scientific | 1284804 | Class II microbiology safety cabinet |
| Scalpel blade number 15 | Fisher Scientific | O305 | |
| Scalpel Swann Morton | Fisher Scientific | 11849002 | |
| Sodium bicarbonate | Sigma | S5761-1KG | |
| Toothed Allis Tissue Forceps | Rocialle | RSPU500-322 | |
| Tryptic Soy Agar | Merck Life Science UK Limited | 14432-500G-F | |
| Tryptic Soy Broth | Merck Life Science UK Limited | 41298-500G-F | |
| Vimoba Tablets | Quip Labs | VMTAB75BX |
References
- Claeys, K. C., et al. Novel application of published risk factors for methicillin-resistant S. aureus in acute bacterial skin and skin structure infections. International Journal of Antimicrobial Agents. 51 (1), 43-46 (2018).
- Rahim, K., et al. Bacterial contribution in chronicity of wounds. Microbial Ecology. 73 (3), 710-721 (2017).
- Guest, J. F., Fuller, G. W., Vowden, P. Costs and outcomes in evaluating management of unhealed surgical wounds in the community in clinical practice in the UK: A cohort study. BMJ Open. 8 (12), 022591 (2018).
- Sen, C. K., et al. Human skin wounds: A major and snowballing threat to public health and the economy. Wound Repair and Regeneration. 17 (6), 763-771 (2009).
- Wilcox, M. H., Dryden, M. Update on the epidemiology of healthcare-acquired bacterial infections: Focus on complicated skin and skin structure infections. Journal of Antimicrobial Chemotherapy. 76, (2021).
- Han, G., Ceilley, R. Chronic wound healing: A review of current management and treatments. Advances in Therapy. 34 (3), 599-610 (2017).
- Percival, S. L., Hill, K. E., Malic, S., Thomas, D. W., Williams, D. W. Antimicrobial tolerance and the significance of persister cells in recalcitrant chronic wound biofilms. Wound Repair and Regeneration. 19 (1), 1-9 (2011).
- Dheman, N., et al. An analysis of antibacterial drug development trends in the United States, 1980-2019. Clinical Infectious Diseases. 73 (11), 4444-4450 (2021).
- MacNeil, S., Shepherd, J., Smith, L. Production of tissue-engineered skin and oral mucosa for clinical and experimental use. Methods in Molecular Biology. 695, 129-153 (2011).
- Yang, Q., et al. Development of a novel ex vivo porcine skin explant model for the assessment of mature bacterial biofilms. Wound Repair and Regeneration. 21 (5), 704-714 (2013).
- Malachowa, N., Kobayashi, S. D., Lovaglio, J., Deleo, F. R. Mouse model of Staphylococcus aureus skin infection. Methods in Molecular Biology. 1031, 109-116 (2013).
- Brandenburg, K. S., Calderon, D. F., Kierski, P. R., Czuprynski, C. J., Mcanulty, J. F. Novel murine model for delayed wound healing using a biological wound dressing with Pseudomonas aeruginosa biofilms. Microbial Pathogenesis. 122, 30-38 (2018).
- Bledsoe, M. J., Grizzle, W. E. The use of human tissues for research: What investigators need to know. Alternatives to Laboratory Animals. , (2022).
- Danso, M. O., Berkers, T., Mieremet, A., Hausil, F., Bouwstra, J. A. An ex vivo human skin model for studying skin barrier repair. Experimental Dermatology. 24 (1), 48-54 (2015).
- Torres, J. P., et al. Ex vivo murine skin model for B. burgdorferi biofilm. Antibiotics. 9 (9), 1-18 (2020).
- Zhao, G., et al. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: A model for the study of chronic wounds. Wound Repair and Regeneration. 18 (5), 467-477 (2010).
- Schierle, C. F., Dela Garza, M., Mustoe, T. A., Galiano, R. D. Staphylococcal biofilms impair wound healing by delaying reepithelialization in a murine cutaneous wound model. Wound Repair and Regeneration. 17 (3), 354-359 (2009).
- Trøstrup, H., et al. Pseudomonas aeruginosa biofilm aggravates skin inflammatory response in BALB/c mice in a novel chronic wound model. Wound Repair and Regeneration. 21 (2), 292-299 (2013).
- Thompson, M. G., et al. Evaluation of gallium citrate formulations against a multidrug-resistant strain of Klebsiella pneumoniae in a murine wound model of infection. Antimicrobial Agents and Chemotherapy. 59 (10), 6484-6493 (2015).
- Maboni, G., et al. A novel 3D skin explant model to study anaerobic bacterial infection. Frontiers in Cellular and Infection Microbiology. 7, 404 (2017).
- Macneil, S. Progress and opportunities for tissue-engineered skin. Nature. 445 (7130), 874-880 (2007).
- Theuretzbacher, U., Outterson, K., Engel, A., Karlén, A. The global preclinical antibacterial pipeline. Nature Reviews Microbiology. 18 (5), 275-285 (2019).
- Miethke, M., et al. Towards the sustainable discovery and development of new antibiotics. Nature Reviews Chemistry. 5 (10), 726-749 (2021).
- Guedes, G. M. M., et al. Ex situ model of biofilm-associated wounds: Providing a host-like environment for the study of Staphylococcus aureus and Pseudomonas aeruginosa biofilms. Journal of Applied Microbiology. 131 (3), 1487-1497 (2021).
- Johnson, C. J., et al. Augmenting the activity of chlorhexidine for decolonization of Candida auris from porcine skin. Journal of Fungi. 7 (10), 804 (2021).
- Horton, M. V., et al. Candida auris Forms High-Burden Biofilms in Skin Niche Conditions and on Porcine Skin. mSphere. 5 (1), 00910-00919 (2020).
- Ashrafi, M., et al. Validation of biofilm formation on human skin wound models and demonstration of clinically translatable bacteria-specific volatile signatures. Scientific Reports. 8, 1-16 (2018).
- Brackman, G., Coenye, T. In vitro and in vivo biofilm wound models and their application. Advances in Experimental Medicine and Biology. 897, 15-32 (2016).
- Rumbaugh, K. P., Carty, N. L. In Vivo Models of Biofilm Infection. Biofilm Infections. , 267-290 (2011).
- Boase, S., Valentine, R., Singhal, D., Tan, L. W., Wormald, P. J. A sheep model to investigate the role of fungal biofilms in sinusitis: Fungal and bacterial synergy. International Forum of Allergy & Rhinology. 1 (5), 340-347 (2011).
- Williams, D. L., et al. Experimental model of biofilm implant-related osteomyelitis to test combination biomaterials using biofilms as initial inocula. Journal of Biomedical Materials Research. Part A. 100 (7), 1888-1900 (2012).
- Scheerlinck, J. P. Y., Snibson, K. J., Bowles, V. M., Sutton, P. Biomedical applications of sheep models: From asthma to vaccines. Trends in Biotechnology. 26 (5), 259-266 (2008).
- Metcalfe, A. D., Ferguson, M. W. J. Tissue engineering of replacement skin: The crossroads of biomaterials, wound healing, embryonic development, stem cells and regeneration. Journal of the Royal Society Interface. 4 (14), 413-417 (2007).
- Kazemi-Darabadi, S., Sarrafzadeh-Rezaei, F., Farshid, A. A., Dalir-Naghadeh, B. Allogenous skin fibroblast transplantation enhances excisional wound healing following alloxan diabetes in sheep, a randomized controlled trial. International Journal of Surgery. 12 (8), 751-756 (2014).
- Martinello, T., et al. Allogeneic mesenchymal stem cells improve the wound healing process of sheep skin. BMC Veterinary Research. 14 (1), 1-9 (2018).
- Roberts, C. D., Windsor, P. A. Innovative pain management solutions in animals may provide improved wound pain reduction during debridement in humans: An opinion informed by veterinary literature. International Wound Journal. 16 (4), 968 (2019).
- Mazzone, L., et al. Bioengineering and in utero transplantation of fetal skin in the sheep model: A crucial step towards clinical application in human fetal spina bifida repair. Journal of Tissue Engineering and Regenerative Medicine. 14 (1), 58-65 (2020).
- Olkowska, E., Gržinić, G. Skin models for dermal exposure assessment of phthalates. Chemosphere. 295, 133909 (2022).
- Couto, N., et al. Label-free quantitative proteomics and substrate-based mass spectrometry imaging of xenobiotic metabolizing enzymes in ex vivo human skin and a human living skin equivalent model. Drug Metabolism and Disposition. 49 (1), 39-52 (2021).

