通过通过纳米孔可调纳米易位流速Zeta电位测定:利用DNA修饰颗粒为例
Summary
在这里,我们使用集成到一个电阻脉冲感测技术经由粒子易位速度的测量,这可以被用来确定各个纳米粒子的ζ电位来表征纳米颗粒表面化学的聚氨酯可调谐纳米孔。
Abstract
纳米孔技术,统称为电阻脉冲传感器(RPS),被用来检测,量化和表征的蛋白质,分子和纳米颗粒。可调电阻脉冲感测(TRPS)是一个相对较新的适应RPS并入,可以实时地改变的可调孔径。这里,我们使用TRPS监测DNA改性的纳米粒子的易位倍,因为它们穿过可调孔膜作为DNA浓度和结构的一个功能( 即 ,单链到双链DNA)。
TRPS是基于两个的Ag / AgCl电极,通过它建立在所施加的电场产生稳定的离子电流的弹性体孔膜分离。与各种基于光学颗粒表征技术,TRPS可以表征单个颗粒之间的样本人口,允许能够轻松进行分析多式联运样品。这里,我们证明zeta电位测量通过已知的标准粒子易位速度和应用这些样品的分析物易位倍,从而导致测量这些分析物的Zeta电位。
以及获得的平均ζ电位的值,将样品全部采用的粒子由粒子立体显示通过样品人口分布的给定样品的详细信息,例如测量的。这样,这种方法演示了医疗和环境等领域的传感应用中的潜力。
Introduction
官能化的纳米颗粒变得如在医疗和环境领域的生物传感器日益流行。的能力,以改变纳米颗粒的表面化学,用DNA,例如,被证明是用于靶向药物递送系统1和监测DNA-蛋白质相互作用2-4是有用的。被利用在生物测定一个越来越普遍的纳米颗粒性质和在治疗剂的递送是超顺磁性5。超顺磁性粒子(SPP的)是在确定和从复杂混合物中去除特定分析物是非常有用的,可以用简单的使用单个磁体的这样做。一旦移除,所述分析物结合的颗粒可以被表征和分析适合目的。
用于纳米颗粒的检测和表征以前的方法包括光学技术,诸如动态光散射(DLS),或称为光子相关光谱法。虽然一喜GH可以通过技术,DLS被限定于一个平均为基础的技术,并没有加入专业软件分析多峰样品时,较大的颗粒将产生一个更主导信号,留下一些较小的颗粒完全被忽视6,7。粒子按颗粒表征技术,因此更利于分析和纳米功能化纳米粒子系统。
基于RPS技术基于周围施加电场到样品,并通过合成或生物纳米孔监测粒子的输送机构。根据RPS相对近期的纳米检测与表征技术是可调电阻脉冲感应(TRPS)8-16。 TRPS是由弹性,可调谐孔隙膜分开的两个电极系统。的可调孔径的方法允许在各种形状17和大小的分析物通过其反待测量通过毛孔口的机制。可调孔隙先前已经用于生产比较的结果,其他技术如透射电子光谱法(TEM)10检测的小颗粒(70-95纳米直径)。当施加电场时,离子电流被观察到并作为颗粒/分子穿过孔,它们暂时阻止孔径,导致在当前的减少,可以被定义为一个“封锁事件'。每个封锁事件是代表单个颗粒,使得样本内的每个颗粒可以表征单独基于所述封锁幅度,Δ 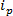 和全宽度半最大,FWHM,以及其他封锁性能。判断单个颗粒,因为他们通过纳米孔为多峰的样品有利的,因为TRPS可以成功地和有效区分的范围内的颗粒尺寸阿蒙GST一个样本。可调电阻脉冲感测完成大小10,同时ζ电位12,18和浓度15测量在单次运行,因此,仍可以区分的类似的样品,如果不通过表面电荷19相同的大小;在施胶替代技术的优势。
和全宽度半最大,FWHM,以及其他封锁性能。判断单个颗粒,因为他们通过纳米孔为多峰的样品有利的,因为TRPS可以成功地和有效区分的范围内的颗粒尺寸阿蒙GST一个样本。可调电阻脉冲感测完成大小10,同时ζ电位12,18和浓度15测量在单次运行,因此,仍可以区分的类似的样品,如果不通过表面电荷19相同的大小;在施胶替代技术的优势。
ζ电位是指在剪切20的平面上的静电势,并作为它们穿过的孔19从粒子速度计算。单个颗粒的ζ电位测量值从而给出洞察易位机制和在溶液中的纳米颗粒系统中,纳米颗粒测定设计未来的有价值的信息用于一系列应用的行为。粒子通过粒子这种性质的分析,还允许之间的样本人群中蔓延,zeta电位值的分布加以探讨,使邻更多信息要达到n个反应动力学(单链到双链DNA,例如)和颗粒稳定性在溶液中。
在这里,我们描述了检测和表征未修饰和DNA修饰SPP表面的技术。本文描述的协议是适用于一系列无机和生物纳米颗粒,但我们证明用DNA修饰表面的方法,由于其广泛的应用。该技术允许用户在纳米颗粒表面的单链和双链DNA靶区分基础上,通过一个孔体系粒子易位速度和因此它们的ζ电势。
Protocol
Representative Results
Discussion
为ζ电势用于计算由Arjmandi 等人 21的工作有关的校准基础的方法。因为它们穿过纳米孔被测量作为所施加电压的函数,使用平均电场和粒子速度定期锥形孔的整体上颗粒的易位持续时间。的电泳迁移率是1 / T(其中T是封锁持续时间)相对于电压的导数,乘以感应区长度的平方, 湖在通过传感区域的多个基准点的平均速度测量以允许最小的误差在使用这种方法计算ζ电位。…
Disclosures
The authors have nothing to disclose.
Acknowledgements
作者感谢IZON科技有限公司的支持。这项工作是由欧洲委员会研究(PCIG11-GA-2012-321836 Nano4Bio)的支持。
Materials
| Phosphate buffered Saline (PBS) | Sigma Aldrich, UK | P4417 | 1 tablet dissolved in 200 mL deionised water to make buffer solution. |
| Tween-20 | Sigma Aldrich, UK | P1379 | 0.05% (v/v) in PBS buffer as a surfactant |
| Carboxyl polystyrene nanoparticles | Bangs Laboratories, US | CPC200 | Nominal diamter of 220 nm, raw concentration of 1E12 particles/mL, specific surface charge of 86 µeq/g (equivalent to a surface charge density of 3.2E19 C/nm^2. |
| Streptavidin coated nanoparticles | Ademtech, France | 3121 | Batch had binding capacity of 4352 pmol/mg (188 nM theoretical DNA binding capacity) at a raw concentration of 1.1E11 particles/mL. |
| Biotinylated oligonucleotides | Sigma Aldrich, UK | VC00001 | Supplier spec: Reverse Phase 1 purification (0.05 Scale); Biotin modification at 3' end; Lyophilised powders reconstituted to 100 µM using deionised water, and diluted as required. Sequences: CP 5'ATGGTTAAACCTCAC TACGCGTGGC[Btn]3' |
| Standard olignonucleotides | Sigma Aldrich, UK | VC00001 | Supplier spec: Reverse Phase 1 purification (0.05 Scale); Lyophilised powders reconstituted to 100 µM using deionised water, and diluted as required. Sequences of DNA targets: Fully complementary – 5'GCCACGCGTAGTGAGGTTTAACCAT3', Middle binding – 5'GTAGTGAGGT3', End binding – 5'GTTTAACCAT3', Partially complementary overhanging – 5'GTGAGGTTTAACCAT TTTTTTTTTTTTTTT3'. |
| Izon qNano | Izon Science, NZ | Inherent pressure on system of 47 Pa, | |
| Izon Variable Pressure Module (VPM) | Izon Science, NZ | Each 'cm' of pressure is equivalent to approximately 1000 Pa. | |
| Polyurethane nanopore membranes | Izon Science, NZ | NP150 | Analyte size range 60-480 nm, pore diameter of calculated to be 799 nm at a 45 mm stretch. |
| Magrack 6 | GE Healthcare, UK | 28-9489-64 | |
| Sonic Bath | Fisher Scientific, UK | 10692353 | 80 Watts |
| Vortexer | IKA, Germany | 0003365000 | |
| Rotary Wheel | Labnet International, US | H5500-230 V |
References
- Alexander, C. M., Maye, M. M., Dabrowiak, J. C. DNA-capped nanoparticles designed for doxorubicin drug delivery. Chem Commun. 47 (12), 3418-3420 (2011).
- Billinge, E. R., Platt, M. Aptamer based dispersion assay using tunable resistive pulse sensing (TRPS). Anal Methods. 7 (20), 8534-8538 (2015).
- Bulyk, M. L. Protein Binding Microarrays for the Characterization of Protein-DNA Interactions. Adv Biochem Eng Biotechnol. 104, 65-85 (2007).
- Platt, M., Rowe, W., Knowles, J., Day, P. J., Kell, D. B. Analysis of aptamer sequence activity relationships. Integr Biol. 1 (1), 116-122 (2009).
- Ruiz-Hernández, E., Baeza, A., Vallet-Regí, M. Smart Drug Delivery through DNA/Magnetic Nanoparticle Gates. ACS Nano. 5 (2), 1259-1266 (2011).
- Murdock, R. C., Braydich-stolle, L., Schrand, A. M., Schlager, J. J., Hussain, S. M. Characterization of Nanomaterial Dispersion in Solution Prior to In Vitro Exposure Using Dynamic Light Scattering Technique. Toxicol Sci. 101 (2), 239-253 (2008).
- Hupfield, S., Holsaeter, A. M., Skar, M., Frantzen, C. B., Brandl, M. Liposome size analysis by dynamic/static light scattering upon size exclusion-/field flow fractionation. J Nanosci Nanotechnol. 6 (7), 3025-3031 (2006).
- Roberts, G. S., et al. Tunable pores for measuring concentrations of synthetic and biological nanoparticle dispersions. Biosens Bioelectron. 31 (1), 17-25 (2012).
- Roberts, G. S., Kozak, D., Anderson, W., Broom, M. F., Vogel, R., Trau, M. Tunable nano/micropores for particle detection and discrimination: scanning ion occlusion spectroscopy. Small. 6 (23), 2653-2658 (2010).
- Vogel, R., et al. Quantitative sizing of nano/microparticles with a tunable elastomeric pore sensor. Anal Chem. 83 (9), 3499-3506 (2011).
- Booth, M. A., Vogel, R., Curran, J. M., Harbison, S., Travas-Sejdic, J. Detection of target-probe oligonucleotide hybridization using synthetic nanopore resistive pulse sensing. Biosens Bioelectron. 45, 136-140 (2013).
- Kozak, D., Anderson, W., Vogel, R., Chen, S. Simultaneous size and ζ-potential measurements of individual nanoparticles in dispersion using size-tunable pore sensors. ACS Nano. 6 (8), 6990-6997 (2012).
- Kozak, D., Anderson, W., Vogel, R., Trau, M. Advances in Resistive Pulse Sensors: Devices bridging the void between molecular and microscopic detection. Nano Today. 6 (5), 531-545 (2011).
- Weatherall, E., Willmott, G. R. Applications of tunable resistive pulse sensing. Analyst. 140, 3318-3334 (2015).
- Willmott, G. R., et al. Use of tunable nanopore blockade rates to investigate colloidal dispersions. J Phys Condens Matter. 22 (45), 454116 (2010).
- Blundell, E. L. C. J., Mayne, L. J., Billinge, E. R., Platt, M. Emergence of tunable resistive pulse sensing as a biosensor. Anal Methods. 7, 7055-7066 (2015).
- Platt, M., Willmott, G. R., Lee, G. U. Resistive Pulse Sensing of Analyte-Induced Multicomponent Rod Aggregation Using Tunable Pores. Small. 8 (15), 2436-2444 (2012).
- Vogel, R., Anderson, W., Eldridge, J., Glossop, B., Willmott, G. A variable pressure method for characterizing nanoparticle surface charge using pore sensors. Anal Chem. 84 (7), 3125-3131 (2012).
- Blundell, E. L. C. J., Vogel, R., Platt, M. Particle-by-Particle Charge Analysis of DNA-Modified Nanoparticles Using Tunable Resistive Pulse Sensing. Langmuir. 32 (4), (2016).
- Hunter, R. J. . Zeta Potential in Colloid Science: Principles and Applications. , (1981).
- Arjmandi, N., Van Roy, W., Lagae, L., Borghs, G. Measuring the electric charge and zeta potential of nanometer-sized objects using pyramidal-shaped nanopores. Anal Chem. 84 (20), 8490-8496 (2012).
- Bacri, L., et al. Dynamics of colloids in single solid-state nanopores. J Phys Chem B. 115 (12), 2890-2898 (2011).
- Cabello-Aguilar, S., et al. Dynamics of polymer nanoparticles through a single artificial nanopore with a high-aspect-ratio. Soft Matter. 10 (42), 8413-8419 (2014).
- Billinge, E. R., Muzard, J., Platt, M. Tunable resistive pulse sensing as a tool to monitor analyte induced particle aggregation. Nanomater Nanosci. 1 (1), 11 (2013).
- Li, J., Fan, C., Pei, H., Shi, J., Huang, Q. Smart Drug Delivery Nanocarriers with Self-Assembled DNA Nanostructures. Adv Mater. 25 (32), 4386-4396 (2013).
- Billinge, E. R., Broom, M., Platt, M. Monitoring aptamer-protein interactions using tunable resistive pulse sensing. Anal Chem. 86 (2), 1030-1037 (2014).
- Gold, L., et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS One. 5 (12), e15004 (2010).
- Park, S. -. J., Taton, T. A., Mirkin, C. A. Array-Based Electrical Detection of DNA with Nanoparticle Probes. Science. 295 (5559), 1503-1506 (2002).

