An Optimized Method for Isolating and Expanding Invariant Natural Killer T Cells from Mouse Spleen
Summary
Here we present an adapted protocol that can be used to generate a large number of murine invariant natural killer T cells from mouse spleen. The protocol outlines an approach by which splenic iNKT cells can be enriched for, isolated and expanded in vitro using a limited number of animals and reagents.
Abstract
The ability to rapidly secrete cytokines upon stimulation is a functional characteristic of the invariant natural killer T (iNKT) cell lineage. iNKT cells are therefore characterized as an innate T cell population capable of activating and steering adaptive immune responses. The development of improved techniques for the culture and expansion of murine iNKT cells facilitates the study of iNKT cell biology in in vitro and in vivo model systems. Here we describe an optimized procedure for the isolation and expansion of murine splenic iNKT cells.
Spleens from C57Bl/6 mice are removed, dissected and strained and the resulting cellular suspension is layered over density gradient media. Following centrifugation, splenic mononuclear cells (MNCs) are collected and CD5-positive (CD5+) lymphocytes are enriched for using magnetic beads. iNKT cells within the CD5+ fraction are subsequently stained with αGalCer-loaded CD1d tetramer and purified by fluorescence activated cell sorting (FACS). FACS sorted iNKT cells are then initially cultured in vitro using a combination of recombinant murine cytokines and plate-bound T cell receptor (TCR) stimuli before being expanded in the presence of murine recombinant IL-7. Using this technique, approximately 108 iNKT cells can be generated within 18-20 days of culture, after which they can be used for functional assays in vitro, or for in vivo transfer experiments in mice.
Introduction
Murine invariant natural killer T (iNKT) cells are a distinct population of innate T lymphocytes selected in the thymus by CD1d-expressing cortical thymocytes 1,2. iNKT cells express a T cell receptor (TCR) comprised of an invariant Vα14-Jα18 TCR chain paired with either Vβ8, Vβ7 or Vβ2 TCRs 3, which is capable of recognizing endogenous as well as foreign lipid antigens in the context of CD1d. For example, murine iNKT cells recognize and are activated by an endogenous lipid antigen called isoglobotrihexosylceramide (iGb3) 4, as well as α-galactosylceramide (αGalCer) 5,6, a glycolipid isolated from marine sponges. TCR-dependent activation of iNKT cells promotes the priming of adaptive immune responses, and as a result, iNKT cells have been shown to be functionally involved in the amelioration or development of a range of pathologies including rheumatic disease 7 and cancer 8. Currently, synthetic iNKT cell ligands constitute promising new vaccine adjuvants that may be capable of regulating a number of immunopathological conditions.
It has previously been demonstrated that iNKT cells can be generated in vitro following isolation from mouse tissue however; many of these studies employ the use of primary antigen-presenting cells (APCs) and/or cell lines 9, Vα14 TCR transgenic (Tg) mice 10, or thymomas for the generation of iNKT cell-derived hybridomas 11,12. Furthermore, large numbers of mice, high volumes of reagents such as αGalCer-loaded CD1d dimers, and lengthy culture times make some published protocols less ethically and economically appealing 9,13.
In this report we describe an adapted method for the isolation and in vitro expansion of iNKT cells from mouse spleen. More specifically, the protocol describes a method for enriching iNKT cells from mouse spleen which reduces the mice, reagents and time required for FACS cell sorting, and proposes an optimized approach for expanding sorted splenic iNKT cells in vitro.
Protocol
In this study, adult (6-8 weeks) female C57Bl/6 mice were used. Mice were housed and bred according to the guidelines of the Ghent University vivarium. All animal procedures were approved by the Institutional Animal Care and Ethics Committee.
1. Preparation of Mononuclear Cells (MNCs) from Mouse Spleen
- Sacrifice the mouse by cervical dislocation.
- Place the mouse back down on the dissecting board and fix the hind and forelegs with pins.
- Sterilize the skin-surface with 70% (vol/vol) ethanol/H2O.
- Cut the skin along the abdominal midline to the thorax and expose the spleen using sterile surgical instruments.
- After trimming the surrounding fat tissues, place the spleen into a 24 well plate containing 1 ml 1x phosphate buffered saline (PBS) at RT.
- Dissect the spleen using a sterile scalpel and transfer the pieces into a 70 µm nylon filter placed within a 50 ml tube.
- Using a 2 ml syringe plunger, gently mash the spleen pieces through the cell strainer and wash with 5 ml 1x PBS.
- Add 3 ml of Ficoll-Paque to a 15 ml tube and overlay the density gradient medium with the 5 ml cellular suspension.
- Centrifuge at 400 x g for 20 min at RT without brake.
- Transfer the interphase to a fresh 15 ml tube, add 10 ml cold 1x PBS and centrifuge at 300 x g for 10 min at 4 °C.
- Resuspend the cells in 2 ml 1x PBS containing 0.5% bovine serum albumin (BSA) and 1 mM ethylenediaminetetraacetic acid (EDTA) (FACS buffer). Transfer 10 µl of the cell suspension to a small tube, mix with 10 µl of 0.4%
2. Enrichment, Detection and Purification of Mouse iNKT Cells
- Enrichment of mouse splenic CD5positive (CD5+) lymphocytes.
- (Optional) Stain 105 cells with FITC-conjugated anti-CD5 (4 µl of 1:30 dilution/106 cells) for 30 min at 4 °C in the dark, wash with 200 µl FACS buffer and resuspend in 500 µl FACS buffer. Prepare a 20 µM working solution of 4',6-diamidino-2-phenylindole (DAPI) in 1x PBS, add 1 µl thereof to 105 cells in FACS buffer and incubate at RT for 5 min. Acquire 104 pre-enriched cells on the flow cytometer.
- Using FCS-H and FCS-W, electronically gate on FSC-Wlo cells to exclude cell doublets and aggregates (Figure 1A). Then electronically gate out DAPI+ (dead) cells within the FSC-Wlo population (Figure 1B), and use SSC-A and FCS-A to electronically select the lymphocyte population by size (Figure 1C).
- Using cells within the lymphocyte gate, plot SSC-A versus CD5 and electronically gate CD5+ lymphocytes as shown in Figure 1D. Acquire the remainder of the pre-enriched sample on the flow cytometer.
- Adjust cell counts to 107 cells per 90 µl in FACS buffer and add 10 µl anti-CD5 microbeads.
- Incubate at 4 ºC for 15 min, wash once with 4 ml FACS buffer and resuspend in 500 µl cold FACS buffer. Enrich for CD5+ lymphocytes using magnetic cell separation (MACS)columns according to manufacturer’s recommendations.
- Optional) Stain 105 cells with FITC-conjugated anti-CD5 (4 µl of 1:30 dilution/106 cells) and DAPI as in 2.1.1. Acquire 104 enriched lymphocytes on the flow cytometer and verify that the electronic gates created in 2.1.1. select CD5+ lymphocytes in the enriched cellular fraction. Acquire 105 cells on the flow cytometer and analyze the percentage CD5+ lymphocytes present in the enriched fraction as shown in Figure 1E–H.
- (Optional) Stain 105 cells with FITC-conjugated anti-CD5 (4 µl of 1:30 dilution/106 cells) for 30 min at 4 °C in the dark, wash with 200 µl FACS buffer and resuspend in 500 µl FACS buffer. Prepare a 20 µM working solution of 4',6-diamidino-2-phenylindole (DAPI) in 1x PBS, add 1 µl thereof to 105 cells in FACS buffer and incubate at RT for 5 min. Acquire 104 pre-enriched cells on the flow cytometer.
- Detection and isolation of mouse iNKT cells by FACS.
- Resuspend enriched CD5+ lymphocytes at a concentration of approximately 106 cells per 20 µl in FACS buffer containing anti-CD16/CD32 (4 µl of 1:50 dilution/106 cells) and PE-conjugated αGalCer/CD1d tetramer (1 µg/106 cells).
- Incubate for 30 min at RT in the dark.
- Dilute mouse antibodies (mAbs) anti-CD3ε V500 (4 µl of 1:20 dilution/106 cells), anti-CD19 e450 (4 µl of 1:50 dilution/106 cells) and anti-CD8α e450 (4 µl of 1:50 dilution/106 cells) in FACS buffer, add to CD5+ lymphocytes and incubate for 30 min at 4 °C in the dark.
- Wash the cells with 1 ml FACS buffer by centrifuging at 300 x g for 10 min at 4 °C and resuspend in 4 ml FACS buffer at a final concentration of 107 cells/ml.
- Filter the cells through a 30 µm cell strainer and place the cells on ice.
- Calibrate the flow cytometer for cell sorting according to the manufacturer’s recommendations 14,15.
Note: Calibration of a flow cytometer for cell sorting requires training and can be performed by an experienced operator, or a trained technician. - Add 10 µl of a 20 µM working solution of DAPI per 106 cells and incubate at RT for 5 min. Acquire approximately 104 enriched CD5+ lymphocytes on the flow cytometer. Electronically gate on FSC-Wlo cells to exclude cell doublets and aggregates (Figure 2A). Then electronically gate out DAPI+ CD8+CD19+ cells (dump channel) within the FSC-Wlo population (Figure 2B), and use SSC-A and FCS-A to electronically select lymphocytes (Figure 2C) as described in 2.1.1.1.
- Plot αGalCer/CD1d tetramer by CD3ε and electronically gate αGalCer/CD1d tetramer+CD3ε+ iNKT cells as shown in Figure 2D. Acquire no more than 2 x 105 CD5+ enriched lymphocytes to determine the percentage αGalCer/CD1d tetramer+CD3ε+ iNKT cells present in the enriched cellular fraction.
- Using the electronic gates created in 2.2.7., FACS sort αGalCer/CD1d tetramer+CD3ε+ iNKT cells on the flow cytometer at 70 psi using a 70 µm nozzle at a flow rate of '1.5'. Collect sorted iNKT cells in a FACS tube containing 1 ml 100% FCS. Subsequently rinse sorted iNKT cells from the side of the FACS tube using 10 ml RMPI 1640 medium and centrifuge the cells at 300 x g for 10 min at 4 °C.
- Resuspend sorted iNKT cells in 1 ml RPMI 1640 medium and acquire 40 µl thereof on a flow cytometer to assess the purity of sorted iNKT cells. Select FSC-Wlo DAPI– lymphocytes as described in 2.2.7. (Figure 2E–G) and electronically gate αGalCer/CD1d tetramer+CD3ε+ iNKT cells as shown in Figure 2H.
3. Culture and Expansion of Mouse iNKT Cells
Day 0
- Coat a flat-bottomed 96 well plate with purified anti-CD3ε (3 µg/ml) per well for 1 hr at RT. Wash twice with 200 µl 1x PBS and once with 200 µl complete RPMI 1640 medium (10% vol/vol FCS, 100 U/ml penicillin–streptomycin, 5.5 µM β-mercaptoethanol).
- Count the number of viable sorted iNKT cells (step 2.2.8) using 0.4% a hemocytometer as in 1.11., centrifuge at 300 x g for 10 min at 4 °C and resuspend at 5 x 105 cells/ml in complete RPMI 1640 medium containing recombinant murine IL-2 (10 ng/ml), recombinant murine IL-12 (1 ng/ml) and soluble anti-mouse CD28 (5 µg/ml). Plate sorted iNKT cells at 105 cells/well in anti-CD3ε coated plates and incubate at 37 °C in a CO2 incubator for 2 days.
Day 2
- Transfer the cells to a fresh uncoated flat-bottomed 96 well plate and culture the cells under resting conditions in complete RPMI 1640 medium at 37 °C in a CO2 incubator for 2 days.
Day 4
- Dislodge the cells from the plate by gentle pipetting and transfer the cells to a fresh 15 ml tube. Centrifuge at 300 x g for 10 min at 4 °C, resuspend at 5 x 105 cells/ml in complete RPMI 1640 medium containing recombinant murine IL-7 (5 ng/ml), and plate 105 cells/well in a flat-bottomed 96 well plate for 4 days at 37 °C in a CO2 incubator.
Day 8
- Collect the cells in a fresh tube, centrifuge at 300 x g for 10 min at 4 °C and resuspend at 5 x 105 cells/ml in complete RPMI 1640 medium containing soluble anti-CD28 (5 µg/ml).
- Coat a flat-bottomed 96 well plate with purified anti-CD3ε (3 µg/ml) per well and wash as in 3.1. Plate 105 cells/well in anti-CD3ε coated plates and incubate at 37 °C in a CO2 incubator until day 11.
Day 11 – 18
- Repeat steps 3.4 to 3.6.
Day 19-20
- Transfer the cells to a fresh 15 ml tube, centrifuge at 300 x g and resuspend at 5 x 105 cells/ml in complete RPMI 1640 medium.
- Plate cells at 105 cells/well in a flat-bottomed 96 well plate and allow the cells to rest for 2 days at 37 °C in a CO2 incubator prior to use in an experiment.
4. Cytokine Production by Mouse iNKT Cells Using a CD1d Plate-bound Assay
- Coat a flat-bottomed 96 well plate with 100 µl of recombinant murine CD1d (10 µg/ml in 1x PBS) per well and incubate for 1 hr at 37 °C.
- Wash four times with 200 µl 1x PBS, add 200 µl of 2% FCS in 1x PBS (blocking solution) per well and incubate for 2 hr at 37 °C.
- Remove the blocking solution, add 200 µl of αGalCer (5 ng/µl) per well, and incubate the plate at 37 °C for 16 hr. [For preparation of the αGalCer solution, add 1,085 µ 1x PBS to 55 µg of αGalCer dissolved in vehicle containing 96 mg/ml sucrose, 10 mg/ml sodium deoxycholate and 5 mg/ml tween 20].
- After incubation, discard the αGalCer solution and wash the well twice with 200 µl 1x PBS followed by 200 µl complete RPMI 1640 medium.
- Collect the in vitro expanded iNKT cells at day 21, centrifuge the cells at 300 x g for 10 min at 4 °C and resuspend in complete RPMI 1640 medium at a concentration of 0.5 x 106 cells/ml.
- Add 200 µl of the iNKT cell resuspension per well and incubate for 72 hr at 37 °C in a CO2 incubator.
- Collect the supernatant and measure cytokines of interest by enzyme-linked immunosorbant assay (ELISA) according to manufacturer’s protocol.
Representative Results
Isolation of splenic mononuclear cells using a density gradient takes approximately 1 hr and eliminates the use of reagents required to lyse red blood cells (RBCs). A high yield of viable cells is obtained using this method and debris generated during straining of the organ is removed. Typically, the frequency of iNKT cells within the splenic lymphocyte pool ranges between 1 and 5% of total T lymphocytes however, this can vary depending on the age, sex and health status of the animals used. Approximately 106 iNKT cells can be acquired from the pooled spleens of 3 mice and the highest yield of iNKT cells are obtained from spleens of 6-8 week old mice.
The use of mouse anti-CD5 magnetic beads significantly enriches for iNKT cells within the splenic MNC fraction and, apart from a minor population of B lymphocytes that express the CD5 antigen, the majority of cells isolated using this procedure constitute CD5+ lymphocytes. For example, 5-8% of the MNCs obtained after CD5 enrichment are CD5-negative (Figure 1H). For iNKT cell FACS sorting, combining αGalCer/CD1d tetramer and anti-mouse CD3ε yields iNKT cell purities above 98% however, FACS dump channels for splenic B and CD8 T cells should be used to eliminate ‘sticky’ lymphocyte populations during the sort. In addition, gate out any doublets that may have formed during the CD5 enrichment step. Using this FACS setup, we obtained iNKT cell purities post-sort in the order of 99% (Figure 2H).
Stimulating 106 sorted iNKT cells with plate-bound anti-CD3ε in the presence of IL-2, IL-12 and soluble anti-CD28 resulted in a 3-fold expansion of iNKT numbers following 2 days of culture (Figure 3). Subsequent expansion iNKT cells in the presence of IL-7 results in a 3- to 4-fold increase in the number of iNKT cells present at day 4 in culture. Notably, repeating these culturing conditions can yield an average of 7 x 107 iNKT cells after two rounds of expansion without any visible loss of cells between changes in culture media on different days (Figure 3). Thus, murine splenic iNKT cells can be expanded at least 70-fold using these conditions in 18 days.
We also assessed the cytokine producing capacity of expanded iNKT cells by stimulation with plate-bound recombinant murine CD1d loaded with αGalCer. At 48 hr post-stimulation, IL-2, IL-4, IFN-γ, TNF-α and IL-6 could be readily detected in culture supernatants by ELISA (Figure 4). Expanded iNKT cells therefore retain the ability to produce Th1 and Th2 cytokines post-expansion.
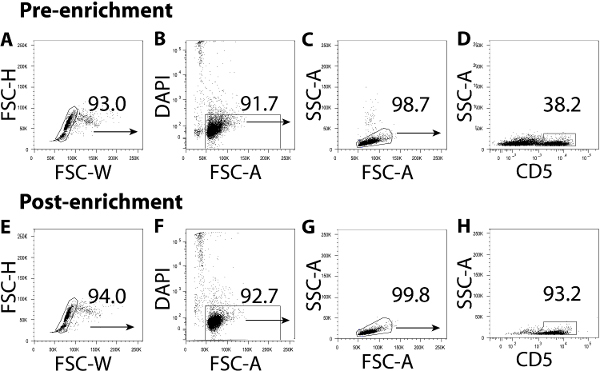
Figure 1. Enrichment for CD5+ lymphocytes from mouse splenic MNCs. Gating strategy for analyzing the percentage of CD5+ lymphocytes present in pre-enriched (A-D) and magnetic cell separation-enriched (E-H) mouse splenic MNC suspensions. Cell doublets (A, E) and dead cells (B, F) have been excluded using FSC-H versus FCS-W and DAPI versus FSC-A plots, respectively. The percentage CD5+ cells present within the lymphocyte gate (C, G) both before and after enrichment are shown in D and H, respectively. Please click here to view a larger version of this figure.
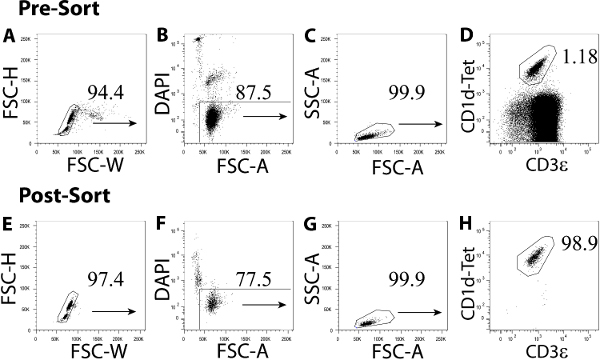
Figure 2. FACS sorting iNKT cells from CD5-enriched splenic MNCs. Gating strategy for analyzing the percentage αGalCer/CD1d tetramer+CD3ε+ iNKT cells present before (A–D) and after (E–H) FACS sorting. The gating strategy eliminates cell doublets (A, E), dead cells and CD8+CD19+ lymphocytes (B, F) from the analysis by plotting FSC-H versus FCS-W and DAPI, CD8 and CD19 (dump channel) versus FSC-A, respectively. The percentage αGalCer/CD1d tetramer+CD3ε+ iNKT cells present within the lymphocyte gate (C, G) before and after FACS sorting are shown in (D) and (H), respectively. CD1d-Tet., αGalCer/CD1d Tetramer. Please click here to view a larger version of this figure.
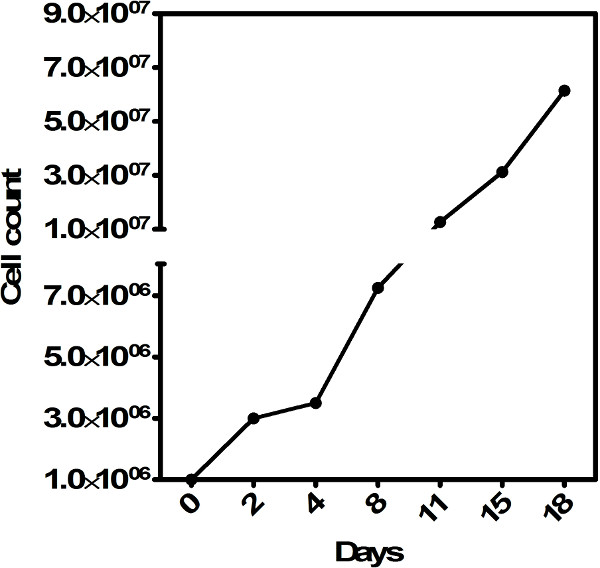
Figure 3. Absolute number of iNKT cells generated in vitro. Fold increase in the number of iNKT cells at the indicated time points after 3 weeks of expansion in vitro. Please click here to view a larger version of this figure.
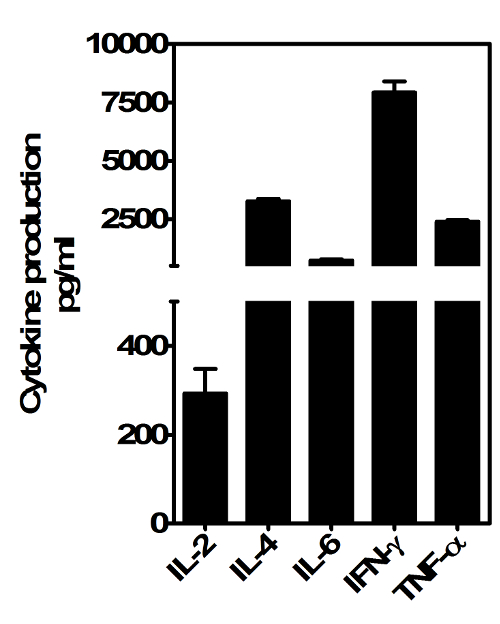
Figure 4. Cytokine producing capacity of in vitro expanded iNKT cells. Day 20 expanded iNKT cells were stimulated in vitro with mouse αGalCer-loaded CD1d. After 72 hr supernatants were collected and analyzed for the presence of IFN-γ, IL-4, TNF-α, IL-2 and IL-6 by ELISA. Bars represent means SEM (n = 2). Please click here to view a larger version of this figure.
Discussion
Critical steps in the current protocol include the isolation and subsequent enrichment of CD5+ lymphocytes (Section 1 & 2), FACS sorting (Section 3) and the initial plating of iNKT cells (Section 4). Of the steps performed in Section 1, remember to carefully layer the splenic cellular suspension over the density gradient medium such that a distinct cellular interphase is generated following centrifugation. The subsequent enrichment for CD5+ lymphocytes by magnetic cell separation (Section 2) minimizes the reagents and time required to stain for and FACS sort iNKT cells (Section 3), which helps increase iNKT cell yields post-sort. Finally, it is important to seed no more than 105 iNKT cells per well for the first 2 days of culture (Section 4) as seeding wells with higher numbers of iNKT cells can result in cellular apoptosis.
A number of considerations are important when modifying and/or troubleshooting this protocol: Firstly, the age of the animals used is important. Animals that are either too old, or have accessory infections, may hamper your ability to isolate, distinguish, and FACS sort sufficient cells from the spleen. Secondly, when staining with αGalCer/CD1d tetramers, it is important to keep the volume of cells resuspended in PBS buffer low as too high a volume dilutes out the staining efficiency of the αGalCer/CD1d tetramer, which in turn reduces the clustering of tetramer+ iNKT cells by FACS. Ideally, the presence of tightly clustered αGalCer/CD1d tetramer+ iNKT cells during FACS acquisition allows for more accurate gating by FACS and yields higher percentage purity iNKTs during post-sort analysis. This is important as lower sorted iNKT cell yields will result in the outgrowth of other contaminating T cell populations during culture. Thirdly, continuously refresh medium that becomes yellow during the culture and expansion of iNKT cells. For example, replace 50% of yellowed medium with new medium containing the appropriate cytokines, particularly during days 8 and day 11 of culture. iNKT cell yields can be maximized in this way.
To our knowledge this is the first iNKT expansion protocol to include a density gradient and CD5+ lymphocyte enrichment step for the isolation and enrichment of iNKT cells from mouse spleen. A number of alternative approaches have been explored to enrich for splenic iNKT cells including αGalCer/CD1d dimers 13, Vα14 TCR Tg mice 10 as well as depletion of non-T cells by magnetic cell separation 9. However, although iNKT cells can be successfully expanded in vitro using these approaches, such methods require either large volumes of αGalCer/CD1d dimers to enrich for iNKTs 13, only generate transgenic iNKT cells 10, or simultaneously give rise to some non-iNKT cell lines 9. Furthermore, the use of plate-bound anti-CD3ε to maintain and expand sorted iNKT cells (Section 4) precludes a requirement for activated APCs during iNKT expansion in vitro 9. In this context, an APC-free iNKT expansion protocol has the advantage of not having to separate APCs from expanded iNKT cells prior to injection in vivo. We should however also add that the protocol described is limited by the fact that the investigator requires easy access to, as well as the relevant training, to calibrate and operate a FACS sorter. Unfortunately FACS sorters are expensive and they are not always available in every laboratory. Nevertheless, although the current protocol has been optimized to isolate and culture murine iNKT cells, the enrichment steps described herein can also be used to isolate and study iNKT recent thymic emigrants (RTEs) 16 and can be adapted to enrich for and characterize other rare T cell populations by FACS.
Collectively, we define an optimized approach that can be used to efficiently generate up to 108 iNKT cells from the spleens of three mice within 3 weeks. iNKT cells generated in this manner retain the ability to secrete cytokines upon stimulation, making them useful for adoptive transfer experiments and the study of iNKT cell biology in vivo.
Disclosures
The authors have nothing to disclose.
Acknowledgements
D.E. and M.B.D. are members of a multidisciplinary research platform (MRP) of Ghent University, and the Ghent Researchers on Unfolded Proteins in Inflammatory Disease (GROUP-ID) consortium.
Materials
| Material | |||
| recombinant murine IL-2 | eBiosciences | 14-8021 | |
| recombinant murine IL-12 | eBiosciences | 39-8122-65 | |
| recombinant murine IL-7 | eBiosciences | 14-8071 | |
| purified anti-mouse CD3e (145-2C11) | eBiosciences | 16-0032-86 | |
| purified anti-mouse CD28 (37.51) | eBiosciences | 16-0281-85 | |
| e450-conjugated anti-mouse CD19 (eBio1D3) | eBiosciences | 48-0193-82 | |
| e450-conjugated anti-mouse CD8α (53-6.7) | eBiosciences | 48-0081-82 | |
| FITC-conjugated anti-mouse CD5 (53-7.3) | eBiosciences | 11-0051-81 | |
| V500-conjugated anti-mouse CD3ε (500A2) | BD biosciences | 560771 | |
| anti-mouse CD5 (Ly-1) microbeads | Macs Miltenyi Biotec | 130-049-301 | |
| purified anti-mouse CD16/32 (2.4G2) | Macs Miltenyi Biotec | 130-092-575 | |
| MACS MS Columns | Macs Miltenyi Biotec | 130-042-201 | |
| Trypan Blue, 0.4% (wt/vol) | Gibco | 15250-061 | |
| RPMI 1640 medium | Gibco | 12633-020 | |
| 1x PBS | Gibco | 10010-056 | [Ca2+/Mg2+ – free] |
| Fetal calf serum | Gibco | 10270 | |
| L-Glutamine | Gibco | 25030-123 | |
| Penicillin-streptomycin | Sigma-Aldrich | P4458-100ML | |
| Bovine serum albumin | Sigma-Aldrich | A7906-100MG | |
| β-Mercaptoethanol | Sigma-Aldrich | M3148 | |
| Ethylenediaminetetraacetic acid | Sigma-Aldrich | EDS-1KG | |
| Ficoll-Paque plus | GE Healthcare | 71-7167-00 AF | |
| Equipment | |||
| 96-well plate F-bottom | Greiner-bio one | 657-160 | |
| 24-well plate | Greiner-bio one | 665-180 | |
| 6-well plate | Greiner-bio one | 655-180 | |
| 15 ml falcon tube | Greiner-bio one | 188-271 | |
| 50 ml falcon tube | Greiner-bio one | 227-261 | |
| 70 µm filter | Greiner-bio one | 542-070 | |
| 30 µm filter | Millipore | SVGP01050 | |
| MiniMACS separator | Macs Miltenyi Biotec | 130-042-102 | |
| MS Columns | Macs Miltenyi Biotec | 130-042-201 | |
| Water-jacketed CO2 incubator | VWR | ||
| Hemocytometer | VWR | ||
| Dissection Kit | VWR | ||
| BD FACSAria III | BD Biosciences |
References
- Bendelac, A., Rivera, M. N., Park, S. H., Roark, J. H. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu Rev Immunol. 15, 535-562 (1997).
- Kronenberg, M., Gapin, L. The unconventional lifestyle of NKT cells. Nat Rev Immuno. 2, 557-568 (2002).
- Park, S. H., et al. The mouse CD1d-restricted repertoire is dominated by a few autoreactive T cell receptor families. J Exp Med. 193, 893-904 (2001).
- Zhou, D., et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 306, 1786-1789 (2004).
- Cardell, S., et al. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J Exp Med. 182, 993-1004 (1995).
- Kawano, T., et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 278, 1626-1629 (1997).
- Drennan, M. B., Aspeslagh, S., Elewaut, D. Invariant natural killer T cells in rheumatic disease: a joint dilemma. Nat Rev Rheumatol. 6, 90-98 (2010).
- Terabe, M., Berzofsky, J. A. NKT cells in immunoregulation of tumor immunity: a new immunoregulatory axis. Trends Immunol. 28, 491-496 (2007).
- Molling, J. W., et al. Generation and sustained expansion of mouse spleen invariant NKT cell lines with preserved cytokine releasing capacity. J Immunol Methods. 322, 70-81 (2007).
- Chiba, A., et al. Rapid and reliable generation of invariant natural killer T-cell lines in vitro. Immunology. 128, 324-333 (2009).
- Gumperz, J. E., et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 12, 211-221 (2000).
- Behar, S. M., Podrebarac, T. A., Roy, C. J., Wang, C. R., Brenner, M. B. Diverse TCRs recognize murine CD1. J Immunol. 162, 161-167 (1999).
- Watarai, H., Nakagawa, R., Omori-Miyake, M., Dashtsoodol, N., Taniguchi, M. Methods for detection, isolation and culture of mouse and human invariant NKT cells. Nature protocols. 3, 70-78 (2008).
- Houtz, B., Trotter, J., Sasaki, D. . BD FACService TECHNOTES. 9 (4), (2004).
- Osborne, G. W. A method of quantifying cell sorting yield in ‘real time’. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 77, 983-989 (2010).
- Drennan, M. B., et al. The thymic microenvironment differentially regulates development and trafficking of invariant NKT cell sublineages. J Immunol. 193, 5960-5972 (2014).

