Using CRISPR/Cas9 to Knock Out GM-CSF in CAR-T Cells
Summary
Here, we present a protocol to genetically edit CAR-T cells via a CRISPR/Cas9 system.
Abstract
Chimeric antigen receptor T (CAR-T) cell therapy is a cutting edge and potentially revolutionary new treatment option for cancer. However, there are significant limitations to its widespread use in the treatment of cancer. These limitations include the development of unique toxicities such as cytokine release syndrome (CRS) and neurotoxicity (NT) and limited expansion, effector functions, and anti-tumor activity in solid tumors. One strategy to enhance CAR-T efficacy and/or control toxicities of CAR-T cells is to edit the genome of the CAR-T cells themselves during CAR-T cell manufacturing. Here, we describe the use of CRISPR/Cas9 gene editing in CAR-T cells via transduction with a lentiviral construct containing a guide RNA to granulocyte macrophage colony-stimulating factor (GM-CSF) and Cas9. As an example, we describe CRISPR/Cas9 mediated knockout of GM-CSF. We have shown that these GM-CSFk/o CAR-T cells effectively produce less GM-CSF while maintaining critical T cell function and result in enhanced anti-tumor activity in vivo compared to wild type CAR-T cells.
Introduction
Chimeric antigen receptor T (CAR-T) cell therapy exhibits great promise in the treatment of cancer.1,2 Two CAR-T cell therapies targeting CD19 (CART19) were recently approved in the United Stated and in Europe for the use in B cell malignancies after demonstrating striking results in multicenter clinical trials.3,4,5 Barriers to more widespread use of CAR-T cells are limited activity in solid tumors and associated toxicities including cytokine release syndrome (CRS) and neurotoxicity (NT).3,5,6,7,8,9 To enhance the therapeutic index of CAR-T cell therapy, genome engineering tools such as zinc finger nucleases, TALENs, and CRISPR are employed to further modify CAR-T cells in an attempt to generate less toxic or more effective CAR-T cells.10,11
In this article, we describe a method to generate CRISPR/Cas9 edited CAR-T cells. The specific goal of this method is to genetically modify CAR-T cells during CAR-T cell manufacturing via CRISPR/Cas9 to generate less toxic or more effective CAR-T cells. The rationale for developing this methodology is built on lessons learned from clinical experience of CAR-T cell therapy, which indicates an urgent need for novel strategies to increase the therapeutic window of CAR-T cell therapy and to extend the application into other tumors and is supported by the recent advances in synthetic biology allowing multiple modifications of CAR-T cells that have started to enter the clinic. While several genome engineering tools are being developed and applied in different settings, such as zinc finger nucleases, TALENs, and CRISPR, our methodology describes CRISPR/Cas9 modification of CAR-T cells.10,11 CRISPR/Cas9 is an RNA-based bacterial defense mechanism that is designed to eliminate foreign DNA. CRISPR relies on endonucleases to cleave a target sequence identified through a guide RNA (gRNA). CRISPR editing of CAR-T cells offers several advantages over other genome engineering tools. These include precision of the gRNA sequence, simplicity to design a gRNA targeting the gene of interest, high gene editing efficiency, and the ability to target multiple genes since multiple gRNAs can be used at the same time.
Specifically in the methods described here, we used a lentivirus encoding CRISPR guide RNA and Cas9 to disrupt a gene during CAR transduction of T cells. In selecting an appropriate technique to edit CAR-T cells, we suggest the technique described here is an efficient mechanism to generate research grade CAR-T cells, but because the long term effect of permanent integration of Cas9 into the genome is unknown, we propose this methodology to develop proof of concept research grade CAR-T cells but not for producing good manufacturing practice grade CAR-T cells.
In particular, here we describe the generation of granulocyte macrophage colony stimulating factor (GM-CSF) knockout CAR-T cells targeting human CD19. These CAR-T cells were generated by transduction with lentiviral particles encoding a guide RNA specific to GM-CSF (gene name CSF2) and Cas9. We previously found that GM-CSF neutralization ameliorates CRS and NT in a xenograft model.12 GM-CSFk/o CAR-T cells allow for the inhibition of GM-CSF during the manufacturing process, effectively reducing production of GM-CSF while enhancing CAR-T cell anti-tumor activity and survival in vivo compared to wildtype CAR-T cells.12 Thus, here we provide a methodology to generate CRISPR/Cas9 edited CAR-T cells.
Protocol
This protocol follows the guidelines of Mayo Clinic's Institutional Review Board (IRB) and Institutional Biosafety Committee (IBC).
1. CART19 cell production
- T cell isolation, stimulation, and ex-vivo culture
- Carry out all cell culture work in a cell culture hood utilizing appropriate personal protective equipment. Harvest peripheral blood mononuclear cells (PBMCs) from de-identified normal donor blood cones collected during apheresis as these are known to be a viable source of PBMCs.13
- To isolate PBMCs, add 15 mL of a density gradient medium to a 50 mL density gradient separation tube. Dilute the donor blood with an equal volume of phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS) to avoid cell trapping.
- Add the diluted donor blood from the cone into the separation tube, careful not to disturb the interface between the blood and the density gradient medium. Centrifuge at 1,200 x g for 10 min at RT.
- Decant the supernatant into a new 50 mL conical, wash with PBS + 2% FBS by bringing up to 40 mL, centrifuge at 300 x g for 8 min at RT, aspirate supernatant, resuspend in 40 mL of buffer, and count cells.
- Isolate T cells from PBMCs via a negative selection magnetic bead kit using a fully automated cell separator according to the manufacturer's protocol.
- To culture the isolated T cells, prepare T cell medium (TCM) that consists of 10% human AB serum (v/v), 1% penicillin-streptomycin-glutamine (v/v), and serum-free hematopoietic cell medium. Sterilize the medium by filtering through a 0.45 µm sterile vacuum filter and then with a 0.22 µm sterile vacuum filter.
- On Day 0 (the day of T cell stimulation), wash CD3/CD28 beads prior to stimulation of T cells. To wash, place the required volume of beads (use enough CD3/CD28 beads for a ratio of 3:1 beads:T cells; note the concentration of beads can be variable) in a sterile 1.5 mL microcentrifuge tube and resuspend in 1 mL of TCM.
- Place in contact with a magnet.
- After 1 min, aspirate the TCM and resuspend in 1 mL of fresh TCM to wash the beads.
- Repeat this procedure for a total of three washes.
- After the third wash, resuspend the beads in 1 mL of TCM.
- Count the T cells.
- Transfer beads to the T cells at a ratio of 3:1 beads:cells.
- Dilute cells to a final concentration of 1 x 106 cells/mL.
- Incubate at 37 °C, 5% CO2 for 24 h.
- T cell transfection and transduction
- Carry out lentiviral work using BSL-2+ precautions, including cell culture hoods, personal protective equipment, and disinfection of used materials with bleach before disposal.
- Acquire a CART19 construct in a lentiviral vector.
NOTE: The CART19 construct utilized here was designed and then synthesized de novo using a commercially available protein synthesis vendor. The CAR construct was subsequently cloned into a third generation lentivirus under control of an EF-1α promoter. The single chain variable region fragment is derived from the FMC63 clone and recognizes human CD19. The CAR19 construct possesses a second generation 4-1BB costimulatory domain and CD3ζ stimulation (FMC63-41BBz).12 - Perform lentiviral production as previously described.12
- In brief, to produce lentivirus, utilize 293T cells that have reached 70-90% confluency.
- Allow incubation for 30 min at room temperature of transfection reagents including 15 µg of the lentiviral plasmid of interest, 18 µg of a gag/pol/tat/rev packaging vector, 7 µg of a VSV-G envelope vector, 111 µL of the pre-complexing reagent, 129 µL of the transfection reagent, and 9.0 mL of the transfection medium before adding to the 293T cells. Then culture the transfected cells at 37 °C, 5% CO2.
- Harvest, centrifuge (900 x g for 10 min), filter (0.45 µM nylon filter), and concentrate supernatant at 24 and 48 h by ultracentrifugation at either 13,028 x g for 18 h or 112,700 x g for 2 h.
- Freeze at -80 °C for future use.
- On Day 1, gently resuspend T cells to break up the rosettes of T cells that had been stimulated at 1 x 106/mL on Day 0.
- Under appropriate BSL-2+ precautions for all lentiviral work, add fresh or frozen harvested virus to the stimulated T cells at a multiplicity of infection (MOI) of 3.0.
- CAR-T cell expansion
- During the phase of expansion, continue to incubate the transduced T cells at 37 °C, 5% CO2. Count CAR-T cells on days 3 and 5, and add fresh, pre-warmed TCM to the culture to maintain a CAR-T cell concentration of 1 x 106/mL.
- Remove beads from the transduced T cells 6 days after stimulation (Day 6) using a magnet. Harvest, resuspend, and place T cells in 50 mL conical tubes in a magnet for 1 min. Then collect the supernatant (contains the CAR-T cells), and discard the beads.
- Place the collected CAR-T cells back in culture at a concentration of 1 x 106 cells/mL to resume expansion.
- On Day 6, assess surface expression of the CAR by flow cytometry.
NOTE: Several methods can be used to detect the surface expression of CAR, such as staining with a goat anti-mouse IgG (H+L) cross-adsorbed secondary antibody or by staining with a CD19 specific peptide, conjugated to a fluorochrome. Here, take an aliquot (about 100,000 T cells) from the culture and wash with flow buffer prepared with Dulbecco's phosphate-buffered saline, 2% fetal bovine serum, and 1% sodium azide. Stain the cells with the anti-CAR antibody and wash twice. Stain the cells with live/dead stain and CD3 monoclonal antibody (OKT3). Wash the cells and resuspend in flow buffer. Acquire on a cytometer to determine transduction efficiency.
- CAR-T cell cryopreservation
- To harvest and cryopreserve CAR-T cells 8 days after stimulation (Day 8 of T cell expansion), harvest the cells from culture.
- Spin down for 5 min at 300 x g.
- Resuspend in freezing medium at 10 million cells per mL per vial in freezing medium consisting of 10% dimethyl sulfoxide and 90% fetal bovine serum.
- Freeze in a freezing container to achieve a rate of cooling of -1 °C/min in a -80 °C freezer and then transfer to liquid nitrogen after 48 h.
- Prior to their use for in vitro or in vivo experiments, thaw CAR-T cells in warm TCM.
- Wash the cells to dilute and remove the dimethyl sulfoxide and resuspend to a concentration of 2 x 106 cells/mL in warm TCM. Rest overnight at 37 °C, 5% CO2.
2. GM-CSF k/o CART19 production
- To disrupt GM-CSF, utilize a guide RNA (gRNA) targeting exon 3 of human GM-CSF (CSF2) selected via screening gRNAs previously reported to have high efficiency for the CSF2 gene that encodes for the human cytokine GM-CSF.14
NOTE: A commercially synthesized research grade Cas9 third generation lentiviral construct containing this gRNA (under a U6 promoter) was used. The construct contains a puromycin resistance gene. The sequence of the gRNA is GACCTGCCTACAGACCCGCC.12 - To produce lentivirus, utilize 293T cells that have reached 70-90% confluency.
- Allow 15 µg of the lentiviral plasmid of interest, 18 µg of a gag/pol/tat/rev packaging vector, 7 µg of a VSV-G envelope vector, 111 µL of the pre-complexing reagent, 129 µL of the transfection reagent, and 9.0 mL of the transfection medium to incubate for 30 min at room temperature.
- Add transfection reagents to the 293T cells. Then culture at 37 °C, 5% CO2.
- Harvest, centrifuge (900 x g for 10 min), filter (0.45 µM nylon filter) and concentrate supernatant at 24 and 48 h by ultracentrifugation at either 13,028 x g for 18 h or 112,700 x g for 2 h and freeze at -80 °C for future use.
- On Day 1, gently resuspend the T cells to break up rosettes.
- In a BSL-2+ approved laboratory, add frozen or freshly harvested virus to the stimulated T cells to generate CAR-T cells. Transduce T cells with both the CAR19 lentivirus and GM-CSF targeting CRISPR lentivirus. Add CAR19 lentivirus at a MOI of 3. Since titration of the CRISPR lentivirus was not feasible, use virus particles generated from a 15 ug plasmid preparation to transduce 10 x 106 T cells.
- See the remaining steps of T cell stimulation, expansion, and cryopreservation as discussed in step 1.
- For lentiCRISPR-edited T cells carrying puromycin resistance, treat cells with puromycin dihydrochloride at a concentration of 1 µg of puromycin per 1 mL on Day 3 and Day 5 .
3. GM-CSF disruption efficiency and functional assessment of GM-CSF k/o CART19
- GM-CSF disruption efficiency: tracking of indels by decomposition (TIDE) sequencing and analysis
- To sequence the exon of interest in GM-CSF gene, use the following primers. Use a forward primer sequence for GM-CSF (CSF2) of TGACTACAGAGAGGCACAGA, and a reverse primer sequence of TCACCTCTGACCTCATTAACC.
- Extract DNA from up to 5 million CAR-T cells with a genomic extraction kit. For the PCR reaction, use 25 µL of a Taq mastermix per reaction with a final concentration of each primer in the PCR reaction of µM, 0.1-0.5 µg of template DNA, and a total volume of the PCR reaction brought up to 50 µL with nuclease free water.
- See the PCR cycling conditions described in Table 1.
- Run PCR samples on a 1-2% agarose gel at 100 V for 30 min and extract using a gel extraction kit.
- Sequence PCR amplicons. Calculate allele modification frequency by uploading raw data into an appropriate TIDE software.15
- GM-CSF production and functional assessment of GM-CSFk/o CART19
- To determine the cytokine production of CAR-T cells, incubate wild type CART19 cells and GM-CSFk/o CART19 with the CD19 expressing acute lymphoblastic leukemia cell line NALM6 at a 1:5 ratio for 4 h at 37 °C, 5% CO2 in the presence of monensin solution, anti-human CD49d, anti-human CD28, and anti-human CD107a.
- Harvest cells from culture for flow cytometry. Wash the cells with flow cytometry buffer. Stain the cells with a live/dead staining kit. Incubate in the dark at room temperature for 15 min. Fix the cells with a fixation medium and incubate in the dark at room temperature for 15 min.
- After washing, stain the cells intracellularly with permeabilization medium in combination with anti-human IFNγ monoclonal antibody, anti-human GM-CSF, anti-human MIP-1β, anti-human IL-2, and anti-human CD3 and incubate in the dark at room temperature for 30 min. Wash and resuspend the samples. Acquire samples on a flow cytometer and analyzed for percentage of intracellular GM-CSF expression.
Representative Results
Figure 1 shows reduction of GM-CSF in GM-CSFk/o CART19 cells. To verify that the genome of the T cells was altered to knockout GM-CSF, TIDE sequencing was used in the GM-CSFk/o CART19 cells (Figure 1A). CAR-T cell surface staining verifies that the T cells successfully express the CAR surface receptor in vitro by gating on live CD3+ cells (Figure 1B). Intracellular staining of GM-CSF by flow cytometry demonstrates decreased expression of GM-CSF in GM-CSFk/o CART19 cells by gating on live CD3+ cells, verifying functional success of the knockout (Figure 1C). GM-CSFk/o CAR-T cells exhibit a decrease in GM-CSF compared to wildtype anti-CD19 CAR-T cells (CART19) via intracellular staining (Figure 1B). In addition, we have previously shown that GM-CSFk/o CAR-T cells reduce production of GM-CSF while enhancing anti-tumor activity and survival compared to wildtype CAR-T cells in vivo.12
| Step | Temp (°C) | Time | Cycles |
| Initial denaturation | 94 | 3 min | 1 |
| Denaturation | 94 | 45 sec | 35 |
| Annealing | 60 | 30 sec | |
| Extension | 72 | 2 min | |
| Post-elongation | 72 | 10 min | 1 |
| 4 | ∞ |
Table 1: PCR cycling conditions for TIDE sequencing of GM-CSFk/o CART19 cells.
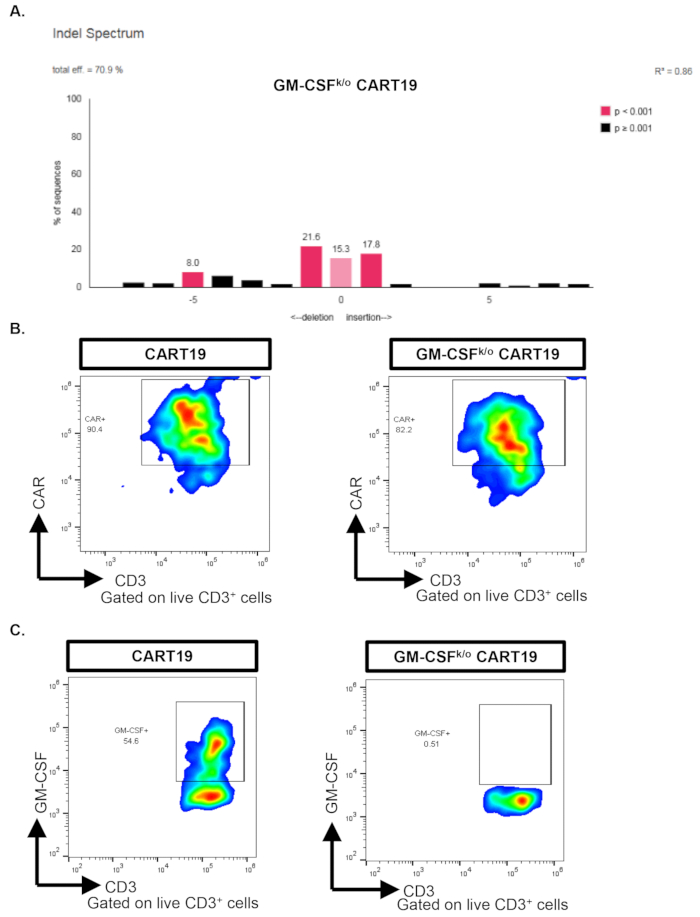
Figure 1: GM-CSFk/o CART19 cells show reduction in GM-CSF.A) A representative TIDE sequence verifies genome alteration of GM-CSF CRISPR/Cas9 GM-CSFk/o CART19 cells with a disruption efficiency of ~71%. B) Representative flow plots depict successful CAR-T cell production with similar CAR expression on wild type CART19 and GM-CSFk/o CART19. C) As assayed by intracellular staining, GM-CSFk/o CART19 cells show reduced GM-CSF compared to wild type CART19 when stimulated with the CD19 positive ALL cell line NALM6. Please click here to view a larger version of this figure.
Discussion
In this report, we describe a methodology to utilize CRISPR/Cas9 technology to induce secondary modifications in CAR-T cells. Specifically, this is demonstrated using lentiviral transduction with a viral vector that contains gRNA targeting the gene of interest and Cas9 to generate GM-CSFk/o CART19 cells. We had previously shown that GM-CSF neutralization ameliorates CRS and NT in a xenograft model.12 As previously described, GM-CSFk/o CAR-T cells allow for the inhibition of GM-CSF during the manufacturing process, effectively reduce production of GM-CSF while maintaining other critical T cell functions, and result in enhanced anti-tumor activity in vivo compared to wildtype CAR-T cells.12
Since permanent integration of Cas9 in the genome could be associated with unwanted effects when translated to a clinically viable product, we propose this specific methodology as a way to generate research grade CRISPR/Cas9 modified CART19 for proof of concept experiments.
While CAR-T cells hold promise in cancer therapy,1,2 their efficacy can continue to be enhanced and their associated toxicities better controlled. CRISPR/Cas9 technology provides a strategy to directly target the genome of CAR-T cells to engineer solutions to current clinical shortcomings. In this article, we describe the generation of GM-CSFk/o CART19 cells.
GM-CSFk/o CAR-T cells were generated by transduction with a lentiviral vector for the CAR-T cell plasmid and a lentivirus construct containing a guide RNA to GM-CSF and Cas9. To generate the GM-CSFk/o, a third generation lentivirus construct (lentiCRISPRv2) containing Cas9 and a reported high efficiency guide RNA (gRNA) targeting exon 3 of human GM-CSF (CSF2)14 under the control of a U6 promoter was utilized. Particular care should be taken during the transduction steps of the procedure as these are the most critical to development of the modified CAR-T cells. Knockout efficiency can be verified and assessed via tracking of indels by decomposition (TIDE) sequences. Functional activity of CAR-T cells is investigated through the antigen specific stimulation of the CAR construct with CD19+ target cells, followed by measurement of different T cell functions, including the production of GM-CSF.12 Of note, staining for CAR expression with a goat anti-mouse antibody should occur before other antibody staining with two washes occurring before surface staining due to the single chain variable region fragment of the CART19 being of mouse origin. This is a point of emphasis in trouble shooting if good CAR expression is not initially observed.
These modified CAR-T cells can be used in in vitro studies and in vivo xenograft models for proof of concept experiments. An advantage of this technique is that CAR-T cell production and genetic manipulation can both occur in one step with good efficiency compared to other techniques.10,11 While dual lentiviral transduction is a convenient and effective method to generate research grade GM-CSFk/o CART19 cells, the DNA is incorporated into the genome and prolonged Cas9 expression may destabilize the genome and increase the risk of off-target effects. A limitation of this technique is that good-manufacturing practice grade knockouts would require modification of the technique to a ribonucleoprotein CRISPR/Cas9 system as this methodology does not result in permanent incorporation of Cas9 into the genome and reduces the potential for off-target effects. The methodology described in the protocol here can potentially be applied to a variety of genes to genetically modify CAR-T cells via CRISPR/Cas9 to help generate less toxic or more effective CAR-T cells.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported through grants from K12CA090628 (SSK), the National Comprehensive Cancer Network (SSK), the Mayo Clinic Center for Individualized Medicine (SSK), the Predolin Foundation (SSK), the Mayo Clinic Office of Translation to Practice (SSK), and the Mayo Clinic Medical Scientist Training Program Robert L. Howell Physician-Scientist Scholarship (RMS).
Materials
| CD3 Monoclonal Antibody (OKT3), PE, eBioscience | Invitrogen | 12-0037-42 | |
| CD3 Monoclonal Antibody (UCHT1), APC, eBioscience | Invitrogen | 17-0038-42 | |
| Choice Taq Blue Mastermix | Denville Scientific | C775Y51 | |
| CTS (Cell Therapy Systems) Dynabeads CD3/CD28 | Gibco | 40203D | |
| CytoFLEX System B4-R2-V2 | Beckman Coulter | C10343 | flow cytometer |
| dimethyl sulfoxide | Millipore Sigma | D2650-100ML | |
| Dulbecco's Phosphate-Buffered Saline | Gibco | 14190-144 | |
| Dynabeads MPC-S (Magnetic Particle Concentrator) | Applied Biosystems | A13346 | |
| Easy 50 EasySep Magnet | STEMCELL Technologies | 18002 | |
| EasySep Human T Cell Isolation Kit | STEMCELL Technologies | 17951 | negative selection magnetic beads; 17951RF includes tips and buffer |
| Fetal bovine serum | Millipore Sigma | F8067 | |
| FITC Mouse Anti-Human CD107a | BD Pharmingen | 555800 | |
| Fixation Medium (Medium A) | Invitrogen | GAS001S100 | |
| GenCRISPR gRNA Construct: Name: CSF2 CRISPR guide RNA 1; Species: Human, Vector: pLentiCRISPR v2; Resistance: Ampicillin; Copy number: High; Plasmid preparation: Standard delivery: 4 μg (Free of charge) |
GenScript | N/A | custom order |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | A-21235 | |
| https://tide.nki.nl. | Desktop Genetics | ||
| Human AB Serum; Male Donors; type AB; US | Corning | 35-060-CI | |
| IFN gamma Monoclonal Antibody (4S.B3), APC-eFluor 780, eBioscience | Invitrogen | 47-7319-42 | |
| Lipofectamine 3000 Transfection Reagent | Invitrogen | L3000075 | |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation | Invitrogen | L34966 | |
| Lymphoprep | STEMCELL Technologies | 07851 | |
| Monensin Solution, 1000X | BioLegend | 420701 | |
| Mouse Anti-Human CD28 Clone CD28.2 | BD Pharmingen | 559770 | |
| Mouse Anti-Human CD49d Clone 9F10 | BD Pharmingen | 561892 | |
| Mouse Anti-Human MIP-1β PE-Cy7 | BD Pharmingen | 560687 | |
| Mr. Frosty Freezing Container | Thermo Scientific | 5100-0001 | |
| NALM6, clone G5 | ATCC | CRL-3273 | acute lymphoblastic leukemia cell line |
| Nuclease Free Water | Promega | P119C | |
| Olympus Vacuum Filter Systems, 500 mL, PES Membrane, 0.22uM, sterile | Genesee Scientific | 25-227 | |
| Olympus Vacuum Filter Systems, 500 mL, PES Membrane, 0.45uM, sterile | Genesee Scientific | 25-228 | |
| Opti-MEM I Reduced-Serum Medium (1X), Liquid | Gibco | 31985-070 | |
| PE-CF594 Mouse Anti-Human IL-2 | BD Horizon | 562384 | |
| Penicillin-Streptomycin-Glutamine (100X), Liquid | Gibco | 10378-016 | |
| Permeabilization Medium (Medium B) | Invitrogen | GAS002S100 | |
| PureLink Genomic DNA Mini Kit | Invitrogen | K182001 | |
| Puromycin Dihydrochloride | MP Biomedicals, Inc. | 0210055210 | |
| QIAquick Gel Extraction Kit | QIAGEN | 28704 | |
| Rat Anti-Human GM-CSF BV421 | BD Horizon | 562930 | |
| RoboSep-S | STEMCELL Technologies | 21000 | Fully Automated Cell Separator |
| SepMate-50 (IVD) | STEMCELL Technologies | 85450 | |
| Sodium Azide, 5% (w/v) | Ricca Chemical | 7144.8-16 | |
| X-VIVO 15 Serum-free Hematopoietic Cell Medium | Lonza | 04-418Q |
References
- Kenderian, S. S., Ruella, M., Gill, S., Kalos, M. Chimeric antigen receptor T-cell therapy to target hematologic malignancies. Cancer Research. 74 (22), 6383-6389 (2014).
- Lim, W. A., June, C. H. The Principles of Engineering Immune Cells to Treat Cancer. Cell. 168 (4), 724-740 (2017).
- Neelapu, S. S., et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. The New England Journal of Medicine. 377 (26), 2531-2544 (2017).
- Maude, S. L., et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. The New England Journal of Medicine. 378 (5), 439-448 (2018).
- Schuster, S. J., et al. Chimeric Antigen Receptor T Cells in Refractory B-Cell Lymphomas. The New England Journal of Medicine. 377 (26), 2545-2554 (2017).
- Grupp, S. A., et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. The New England Journal of Medicine. 368 (16), 1509-1518 (2013).
- Porter, D. L., Levine, B. L., Kalos, M., Bagg, A., June, C. H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. The New England Journal of Medicine. 365 (8), 725-733 (2011).
- Gust, J., et al. Endothelial Activation and Blood-Brain Barrier Disruption in Neurotoxicity after Adoptive Immunotherapy with CD19 CAR-T Cells. Cancer Discovery. 7 (12), 1404-1419 (2017).
- Fitzgerald, J. C., et al. Cytokine Release Syndrome After Chimeric Antigen Receptor T Cell Therapy for Acute Lymphoblastic Leukemia. Critical Care Medicine. 45 (2), e124-e131 (2017).
- Liu, X., Zhao, Y. CRISPR/Cas9 genome editing: Fueling the revolution in cancer immunotherapy. Current Research in Translational Medicine. 66 (2), 39-42 (2018).
- Ren, J., Zhao, Y. Advancing chimeric antigen receptor T cell therapy with CRISPR/Cas9. Protein Cell. 8 (9), 634-643 (2017).
- Sterner, R. M., et al. GM-CSF inhibition reduces cytokine release syndrome and neuroinflammation but enhances CAR-T cell function in xenografts. Blood. , (2018).
- Dietz, A. B., et al. A novel source of viable peripheral blood mononuclear cells from leukoreduction system chambers. Transfusion. 46 (12), 2083-2089 (2006).
- Sanjana, N. E., Shalem, O., Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods. 11 (8), 783-784 (2014).
- Brinkman, E. K., Chen, T., Amendola, M., van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Research. 42 (22), e168 (2014).

