Microinjection of DNA into Eyebuds in Xenopus laevis Embryos and Imaging of GFP Expressing Optic Axonal Arbors in Intact, Living Xenopus Tadpoles
Summary
This protocol aims to demonstrate how to microinject a DNA/DOTAP mixture into eyebuds of one day old Xenopus laevis embryos, and how to image and reconstruct individual green fluorescent protein (GFP) expressing optic axonal arbors in tectal midbrains of intact, living Xenopus tadpoles.
Abstract
The primary visual projection of tadpoles of the aquatic frog Xenopus laevis serves as an excellent model system for studying mechanisms that regulate the development of neuronal connectivity. During establishment of the retino-tectal projection, optic axons extend from the eye and navigate through distinct regions of the brain to reach their target tissue, the optic tectum. Once optic axons enter the tectum, they elaborate terminal arbors that function to increase the number of synaptic connections they can make with target interneurons in the tectum. Here, we describe a method to express DNA encoding green fluorescent protein (GFP), and gain- and loss-of-function gene constructs, in optic neurons (retinal ganglion cells) in Xenopus embryos. We explain how to microinject a combined DNA/lipofection reagent into eyebuds of one day old embryos such that exogenous genes are expressed in single or small numbers of optic neurons. By tagging genes with GFP or co-injecting with a GFP plasmid, terminal axonal arbors of individual optic neurons with altered molecular signaling can be imaged directly in brains of intact, living Xenopus tadpoles several days later, and their morphology can be quantified. This protocol allows for determination of cell-autonomous molecular mechanisms that underlie the development of optic axon arborization in vivo.
Introduction
During development of the nervous system, axons of presynaptic neurons navigate through diverse regions of the brain to reach their target areas. When axons invade their target tissues, they establish synaptic connections with postsynaptic target neurons. In many types of neurons, axons increase the number and spatial extent of synaptic connections they can make by elaborating networks of terminal branches or arbors1. The retino-tectal projection of tadpoles of the aquatic frog Xenopus laevis is a powerful vertebrate model for examining mechanisms underlying terminal axon arborization and synaptic connectivity2,3,4. Individual GFP expressing optic axonal arbors with normal and altered molecular signaling can be observed directly in intact, living Xenopus tadpoles5,6,7,8. To express GFP alone or together with full-length or truncated versions of genes in small number of optic neurons, we use a technique involving microinjection/lipofection of DNA into eyebuds of one day old Xenopus embryos9,10. This technique was originally developed to study mechanisms of optic axon pathfinding in young Xenopus tadpoles, and has since been applied by us and others to determine cell-autonomous molecular mechanisms underlying optic axon arborization in Xenopus tadpoles5,6,7,8,9,10.
Alternate techniques to express exogenous genes in a small number of optic neurons have been developed in other model species, as well as in X. laevis. However, each of these approaches presents challenges and limitations when compared to microinjection of DNA/lipofection reagent in eyebuds of Xenopus embryos. In mice, transgenesis can be used to express genes in a small number of optic neurons, but the generation of transgenic mice is costly and time consuming and transgenic mice often present with undesirable side effects11. Transgenic zebrafish that express exogenous genes in optic neurons can also be created by injecting plasmids into early cleavage stage embryos12. However, this process requires cloning of a specific promoter to express genes in a mosaic pattern in optic neurons in zebrafish larvae12. The frequency of expression of exogenous DNA in optic neurons in transgenic zebrafish is also somewhat lower (<30%) compared to Xenopus tadpoles that were microinjected with DNA/liposomal reagent (30−60%)12. In ovo electroporation has also been used to express genes in small numbers of optic neurons in chicks13. However, this procedure has failed to fully characterize mechanisms that establish optic projections because optic axon arborization cannot be imaged in intact, living chick embryos. Finally, several laboratories have used electroporation to transfect genes into small number of optic neurons in Xenopus tadpoles14,15. Yet, electroporation requires optimization of equipment and protocols (stimulator, electrodes, spatial and temporal patterns of wave pulses) beyond that used for microinjection of DNA/lipofection reagent into eyebuds of Xenopus embryos.
We and others previously used the technique of microinjection/lipofection of DNA into eyebuds of Xenopus embryos to determine cell autonomous signaling mechanisms that establish optic axon arborization5,6,7,8. We initially used this approach to dissect the functions of the Cadherin and Wnt adaptor protein β-catenin in optic axonal arborization in Xenopus tadpoles5,6. In one study, we showed that β-catenin binding to α-catenin and to PDZ is required, respectively, for initiating and shaping optic axonal arbors in vivo5. In a second report, we demonstrated that the β-catenin binding domains for α-catenin and GSK-3β oppositely modulate projection patterns of ventral optic axonal arbors6. More recently, we identified roles for the Wnt factor, adenomatous poliposis coli (APC), in regulating morphological features of optic axonal arbors in Xenopus tadpoles7. By co-expressing the N-terminal and central domains of APC that modulate β-catenin stability and microtubule organization together with GFP in individual optic neurons, we determined shared and distinct roles for these APC interaction domains on branch number, length, and angle in optic axonal arbors in vivo7. Another laboratory used the microinjection/lipofection technique to determine cell autonomous roles for signaling by the BDNF receptor, TrkB, in optic axonal arbors in Xenopus tadpoles8. This group showed that expression of a dominant-negative TrkB perturbed branching and synaptic maturation in individual optic axon arbors in vivo8. Overall, the lipofection technique in Xenopus has already illuminated the specific roles of different genes in optic axon branching in the native environment.
Protocol
All methods described here have been approved by the Institutional Animal Care and Use Committee (IACUC) of Touro University California (Protocol # TUCA003TE01X).
1. Obtaining X. laevis Embryos
- Obtain X. laevis embryos by natural mating of pairs of male and female adult frogs primed with human chorionic gonadotropin (HCG), by in vitro fertilization of eggs shed from female adult frogs primed with HCG, or by ordering directly (Table of Materials).
- Dejelly embryos obtained with a 2% cysteine solution at room temperature (Table of Materials)16.
- Collect 50−100 embryos in a large Petri dish. Remove the solution the embryos are in by decanting or using a plastic transfer pipette. Add 25 mL of 2% cysteine solution (0.5 g cysteine in 25 mL ddH2O, pH to 8.0) to the dish containing the embryos.
- Gently swirl the Petri dish containing the embryos in the cysteine solution until the jelly coats of the embryos fall off and the embryos collect in a clump in the center of the dish (5−10 min). At this point, slowly and gently pour off the cysteine solution into a waste beaker. Take care not to pour too many of the embryos in the waste beaker along with the cysteine solution.
- Rinse the embryos in the Petri dish 6x with 10% modified Mark’s Ringer solution (MMR) or other suitable solution (e.g., modified Barth’s Solution, MBS), swirling the dish each time the solution is replaced.
- Culture the embryos in 10% MMR until they reach developmental stages 22−2417. Xenopus embryos can be incubated at temperatures between 15−25 °C. The rate of development of the embryos depends on the temperature they are incubated at17.
NOTE: Xenopus embryos ordered from a catalogue usually arrive in the laboratory at developmental stages 20−24, so they can be dejellied and microinjected right away.
2. Preparing DNA Plasmids and Making a DNA/DOTAP Mixture
- Subclone DNA expression constructs into Xenopus expression vectors pCS2+ or pCS2+MT or derivatives thereof (originally constructed by D. Turner and R. Rupp)5,6,7. pCS2+ vectors contain a modified cytomegalovirus (CMV) promoter that facilitates gene expression in frogs.
- Amplify pCS2 plasmids containing GFP and/or genes of interest with miniprep kits (Table of Materials) following the standard procedure. In the final elution step of the miniprep protocol, perform a sequential elution of the DNA into ddH2O to yield a final concentration of >1 µg/µL.
- Store all pCS2 plasmids at -80 °C until ready to perform a microinjection/lipofection experiment, i.e., when embryos are at developmental stages 22−24.
- Thaw DNA plasmids to be lipofected at room temperature. Immediately prior to lipofection, briefly centrifuge DNA plasmids. This will prevent precipitate from forming in the DNA/DOTAP mixture that could clog the tip of the microcapillary pipette.
- Combine DNA plasmids with the DOTAP liposomal transfection reagent (Table of Materials) at a 1:3 (w/v) ratio9,10. For example, transfer 2 µg of DNA to a 1.5 mL microcentrifuge tube and add 6 µL of DOTAP, or transfer 3 µg of DNA in a microcentrifuge tube and add 9 µL of DOTAP.
- Once the DNA and DOTAP are combined, gently flick the microcentrifuge tube to mix the solution. The DNA/DOTAP solution should become slightly opaque after mixing.
- If two plasmids are to be lipofected together (e.g., pCS2-GFP with a second pCS2 plasmid containing a truncated or full-length version of a gene) in optic neurons, first combine the two plasmids (after briefly centrifuging both of them) at a 1:1 ratio, and then add DOTAP at a 1:3 (w/v) ratio. For example, combine 1 µg of pCS2-GFP with 1 µg of a second pCS2 plasmid and then add 6 µL of DOTAP.
NOTE: Studies have shown that lipofection of two plasmids into eyebuds of Xenopus embryos at these developmental stages will result in their co-expression in individual optic neurons9,10.
3. Loading a Microinjection Needle with DNA/DOTAP
- Gently clip the tip of a pulled glass microcapillary pipette with fine forceps (Table of Materials).
- Backfill the glass microcapillary pipette with mineral oil using a microfil such that a tiny drop of mineral oil appears at the clipped tip of the micropipette. Fill the microcapillary pipette halfway with mineral oil.
- Load the pulled glass microcapillary pipette that is now filled with mineral oil into a suitable injection holder connected to an injector. If using an injector (Table of Materials), eject the plunger halfway before loading the microcapillary pipette onto it. Once the microcapillary pipette is securely attached to the injector, extend the plunger to the full extent to confirm that the microcapillary pipette is strongly attached to the injector and does not move with the extension of the plunger.
- Transfer a 3 μL drop of the DNA/DOTAP mixture onto a cut square (1 inch square) sheet of paraffin paper.
- Under a stereo dissecting microscope, move the tip of the glass microcapillary pipette into the DNA/DOTAP drop.
- Slowly suck the DNA/DOTAP drop into the glass microcapillary pipette using the fill option on the injection apparatus. As the liquid is loaded into the microcapillary pipette, the drop will get smaller. Due to the slight opacity of the DNA/DOTAP solution, the boundary between the mineral oil and the DNA/DOTAP solution should be visible in the glass microcapillary pipette (Figure 1). If needed, stop filling the microcapillary pipette periodically to allow the pressure in the glass microcapillary pipette to recalibrate.
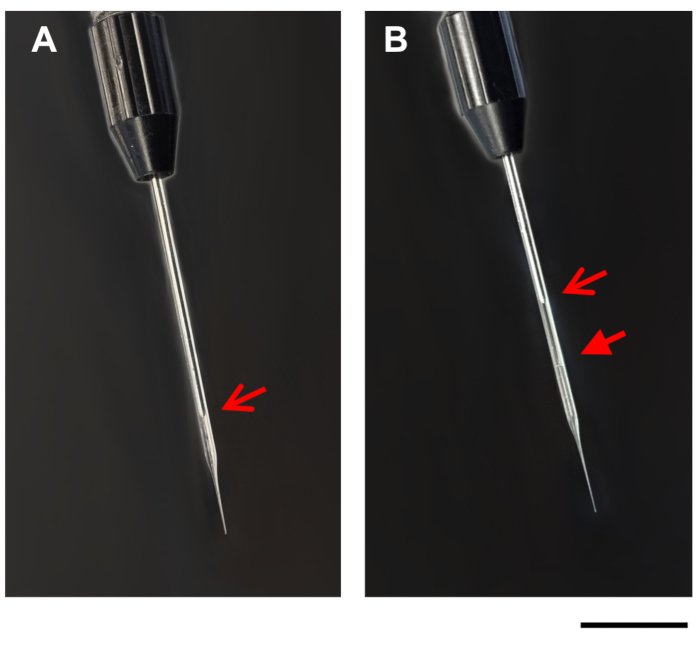
Figure 1: Images of microcapillary pipette. Images show a microcapillary pipette on the injection apparatus, before (A), and after (B) filling with DNA/DOTAP. Open arrows, tip of plunger in the microcapillary pipette (A,B). Closed arrow, line between mineral oil and DNA/DOTAP in the filled microcapillary pipette (B). Scale bar = 1 mm. Please click here to view a larger version of this figure.
4. Microinjecting DNA/DOTAP into Eyebuds of 1 Day Old Xenopus Embryos
- Manually devitellinize ten stage 20−24 Xenopus embryos with fine forceps in a 10 mm Petri dish filled with 0.1x MMR. Grasp the vitelline envelope at the waist to avoid injuring the embryos. With forceps in both the experimenter’s left and right hands, pop the bubble of the vitelline envelope and release the embryo from the vitelline envelope. Take care not to injure the embryos when removing the vitelline envelope.
NOTE: Beginning at stage 20, Xenopus embryos develop an indentation or ‘waist’ between the anterior and posterior halves of the embryo. This waist allows a gap to form between the vitelline envelope and the embryo at this position. - Use a plastic transfer pipette with a cut tip to transfer 5−10 devitellinized stage 22−24 embryos to a 10 mm Petri dish filled with 1x MMR.
NOTE: The higher salt solution in 1x MMR facilitates healing of puncture wounds that will result from the microinjection. - Under the stereomicroscope, grasp one of the devitellinized embryos in the Petri dish with forceps in each hand, and arrange the embryo so that its anterior pole is pointed up in the field of view. Orient the embryo so that it is lying laterally and one of its eyebuds (left or right) is facing upwards.
- Under the stereomicroscope, hold the embryo with the forceps in the experimenter’s non-dominant hand, and with the experimenter’s dominant hand introduce the tip of the glass micropipette into the eyebud (from the ventral or the dorsal side, depending on the half of the embryo that is currently being injected), just beneath the epidermis (Figure 2). Inject between 70−210 nL of the DNA/DOTAP solution. This can be done in several pulses, depending on the size of the pulse the injector is set to (usually 70 nL).
NOTE: The deepness of injection is very important for lipofection into optic neurons. If the position of the tip of the microcapillary is correctly inserted very superficially into the eyebud, then following microinjection, the grey epidermis overlying the eyebud will swell. If the position is too deep, the grey eyebud will not show any changes, and the frequency of expression of DNA in optic neurons will be lower. - Turn the embryo around and perform the same microinjection into the eyebud on the contralateral side of the embryo.
- Inject both eyebuds of 6−10 (or more) embryos in each experiment.
- After microinjection, store embryos in a Petri dish with 1x MMR for approximately 30 min to facilitate wound healing.
- After 30 min, transfer injected embryos into a 0.1x MMR solution with 0.001% bleaching agent (phenylthiocarbamide) to reduce pigmentation. Culture the embryos covered for approximately five days, until the embryos have developed into tadpoles at stages 46−4716.
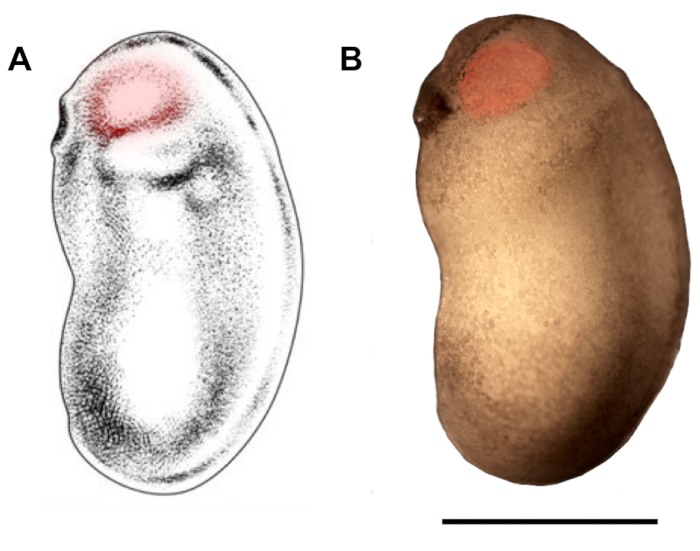
Figure 2: Demarcation of eyebud region for microinjection. Schematic (A) and photomicrograph (B) of X. laevis embryo at developmental stages 22/23 show eyebud region that should be targeted for microinjection (red highlights). Scale bar = 1 mm. Panel A has been modified from Zahn et al.18. Please click here to view a larger version of this figure.
5. Imaging of GFP Expressing Optic Axonal Arbors in Intact, Living Tadpoles
NOTE: When tadpoles that were lipofected with DNA reach developmental stages 46−47, they are ready for imaging.
- Prior to imaging, tadpoles must be anesthetized. To anesthetize tadpoles, transfer the lipofected tadpoles into a 0.02% tricaine solution in ddH2O in a 10 mm Petri dish. Wait 5−10 min until tadpoles become immobile. Verify that tadpoles are still alive by observing their beating hearts under a stereo dissecting microscope.
- Place one anesthetized tadpole into a custom-made silicone chamber on a glass slide and seal with a coverslip. The tadpole should tilt slightly so that one side (left or right) of its head is angled upwards and just barely touches the cover slip.
- Screen the half of the dorsal tectal midbrain of the tadpole that is tilted upwards at low magnification for GFP expressing optic axonal arbors.
NOTE: A widefield upright microscope equipped with an epilfuorescence illumination and an apochromatic objective lens (Table of Materials) can be used to screen for fluorescent arbors. - If the tectal hemisphere contains between one to three GFP expressing optic axonal arbors, capture a z-series of images of these arbors using a high contrast 40x air long working distance objective (Table of Materials). For each axonal arbor, capture 10−20 z-series slices at 1.5 µm intervals.
- In order to view axon arbors on the other side of the tectal midbrain, reload the tadpole in the silicon chamber so that it tilts to the other side and seal with a cover slip. Then repeat steps 5.3 and 5.4.
6. Reconstruction and Quantification of Optic Axonal Arbor Morphology
- Select an image stack that contains between one to three GFP expressing optic axonal arbors.
- Use the freehand drawing tool in graphic editing software (Table of Materials) to trace the portion of each optic axonal arbor visible in each z-slice. Tracing through the pieces of each arbor evident in each z-slice will create an accurate 2D projection of the arbor. Different colors can be used to trace distinct GFP expressing optic axonal arbors.
- Make all morphometric measurements on 2D reconstructions of the axonal arbors, with reference to the original z-series of images when needed7. Using Image J software (Table of Materials), measure morphological parameters such as number of branches (i.e., number of branch tips or branch points), total arbor branch length, length per branch, length and width of arbor, overall shape of arbor (L/W ratio, circularity), and angle of branches7.
Representative Results
The protocol described in this article yields a success rate of 30−60% of injected Xenopus embryos expressing GFP (alone or together with an additional DNA constructs) in one to ten optic axonal arbors. In Figure 3, we show representative confocal images of GFP expressing control and mutant optic axonal arbors in intact Xenopus tadpoles from our recently published study7. For this study, we cloned two domain mutants of APC (APCNTERM and APCβ-cat) into pCS2 plasmids, and co-injected these plasmids together with a pCS2-GFP labelling plasmid into eyebuds of one day old Xenopus embryos. Figure 4 shows results of several quantitative measurements we made on reconstructions of the control and APC mutant axonal arbors, including number of branches, total arbor branch length, and mean length of branches.
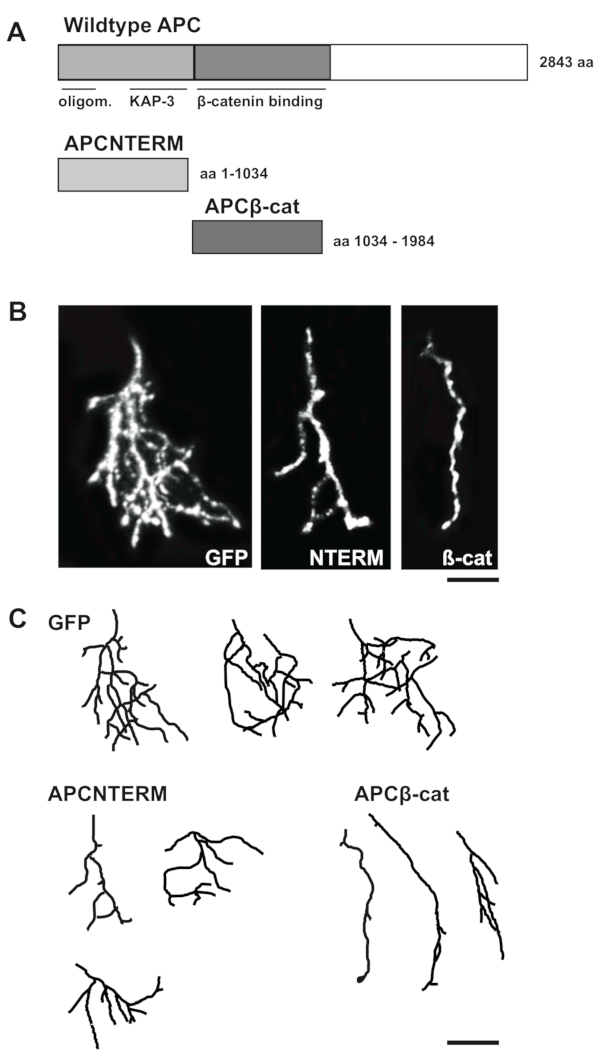
Figure 3: Representative images of GFP expressing control and mutant optic axonal arbors. (A) Schematic of mutants of APC N-terminal and central domain mutants that were cloned into pCS2 plasmids. (B) Representative confocal images of single GFP and GFP-APC mutant optic axonal arbors in tecta of intact, living Xenopus tadpoles. (C) Reconstructions of z-series images of GFP control and APC mutant optic axonal arbors. Scale bars = 30 µm (B), 40 µm (C). This figure has been modified from Jin et al.7. Please click here to view a larger version of this figure.
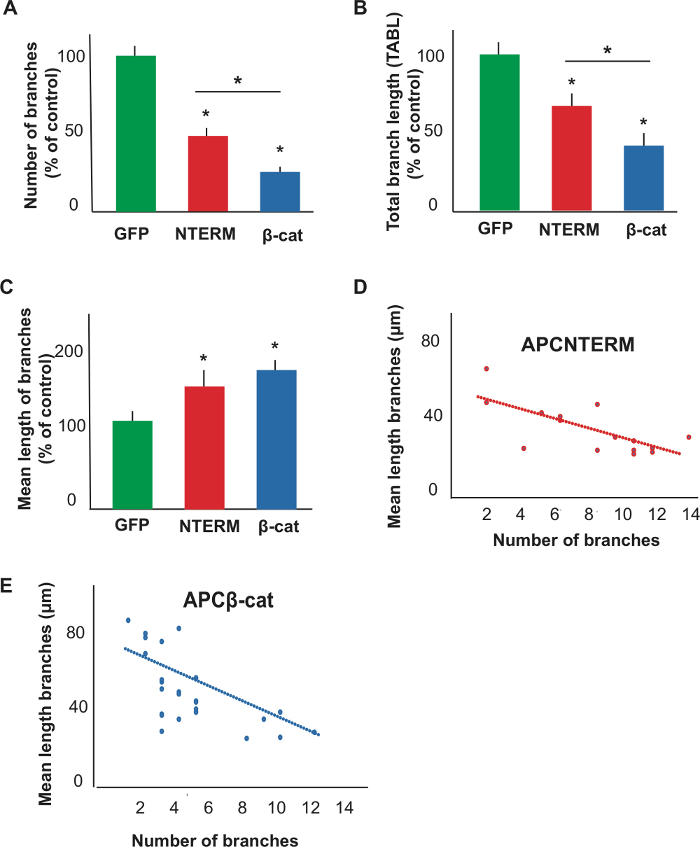
Figure 4: Quantification of morphologies of reconstructions of control and mutant axonal arbors. Plots of number of branches (A), total arbor branch length (B), and mean branch length (C) confirm observed differences between control and APC mutant expressing axonal arbors. Data in panels A−C is shown as percent of control mean with SEM. *above data bar or line indicates p < 0.05. Additional scatter plots of number of branches versus mean branch length with regression lines show inverse correlation between these parameters in optic axonal arbors expressing APC domains (D, E). Sample numbers: (A) GFP-12, APCNTERM–18 APCβ-cat-25; (B) GFP-12, APCNTERM-16, APCβ-cat-25; (C) GFP-11, APCNTERM-16, APCβ-cat-25. This figure has been modified from Jin et al.7. Please click here to view a larger version of this figure.
Discussion
In this article, we demonstrate how to express exogenous DNA constructs in single or small numbers of optic neurons and how to image individual GFP expressing optic axonal arbors with normal and altered molecular signaling in intact, living tadpoles of the frog X. laevis. We also explain how to reconstruct and quantify the morphology of GFP expressing optic axonal arbors from images captured in vivo. To express exogenous DNA plasmids in small number of optic neurons, we microinject a DNA/lipofection reagent mixture into eyebud primordia of one day old Xenopus embryos, using a technique first developed in the laboratory of Christine Holt to study optic axon pathfinding in young tadpoles9,10,19,20,21. We and others have also applied this DNA microinjection/lipofection technique to study molecular mechanisms that regulate optic axon arborization in older intact, living X. laevis tadpoles5,6,7,8. This inexpensive, simple procedure for transient, cell specific transgenesis allows determination of cell-autonomous gene function in developing optic axonal arbors in a living vertebrate model system.
There are several important factors to consider for best practices when microinjecting DNA/DOTAP into eyebuds of Xenopus embryos. First, as noted in other reports, the DNA concentration should be greater than 1 µg/µL9,10. DNA concentrations between 1−3 µg/µL are best, but DNA concentrations as low as 0.7 µg/µL can also be used. Second, microinjecting DNA into stage 22−24 Xenopus embryos is optimal for experiments in which the goal is to examine mechanisms regulating optic axonal arborization. In embryos at these developmental stages, eyebuds are morphologically differentiated and can be more easily targeted for injection14. Most optic neurons in the eyebud primordia of stage 22−24 embryos are also post-mitotic, which results in a smaller number of optic neurons expressing GFP, which in turn allows for better resolution when imaging individual GFP optic axonal arbors in tadpoles. Finally, previous studies have shown that exogenous genes are expressed ~8 h after lipofection9. Therefore, expressing gene constructs in eyebuds of stage 22−24 embryos means that the genes will perturb neither cell fate selection of optic neurons nor the initial outgrowth of their axons. A third factor that will ensure success when microinjecting DNA into Xenopus embryo eyebuds is that the DNA should be injected into a relatively superficial region of the eyebud9,10. Injecting into more deep tissues in or around the eyebud will result in a lower percentage of embryos that contain optic neurons expressing the exogenous DNA10.
There are also several issues to be aware of when imaging and reconstructing GFP expressing optic axonal arbors in intact, living Xenopus tadpoles. First, researchers should only capture images of tectal hemispheres that contain between one to three GFP expressing optic axonal arbors. If a single image contains more than three GFP expressing optic axonal arbors, the arbors are likely to have overlapping branches, which will make it difficult to define the individual arbors during the reconstruction process. Another issue to be aware of is that the reconstruction and quantification of optic axonal arbor morphology are the most time-consuming steps in this protocol. We estimate that reconstructing and quantifying optic axonal arbor morphology require approximately 80−90% of the total time of the experiment. Although techniques have been developed to automate the reconstruction of tectal neuron dendritic arbors, these computational methods have not yet been applied to optic axonal arbors as well22. Although laborious, the process of reconstructing optic arbor morphology dramatically increases researchers’ comprehension of the details of optic axonal arbor morphology. This added detail, in turn, significantly improves the quality of images of GFP expressing arbors these researchers are able to capture in future experiments.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Touro University California College of Osteopathic Medicine for supporting our research. We acknowledge previous students in the laboratory (Esther Wu, Gregory Peng, Taegun Jin, John Lim) who helped implement this microinjection technique in our laboratory. We are grateful to Dr. Christine Holt, in whose laboratory this DNA microinjection/lipofection technique in Xenopus embryos was first developed.
Materials
| 3.5" Micropipettes | Drummond Scientific | 3-000-203 – G/X | |
| μ-manager software (Version ) | www.micro-manager.org | ||
| CCD camera | Scion Corporation | CFW-1312 M | |
| Chorulon (Human Chorionic Gonadotropin) | AtoZ Vet Supply | N/A | |
| Cysteine | Sigma-Aldrich | 168149-100G | |
| DOTAP | Sigma-Aldrich | 11202375001 | |
| Dumont Forceps #5 | Fine Science Tools | 11250-10 | |
| Eclipse E800 epifluoresence microscope | Nikon | Objectives: Nikon Plan Apo 20X/0.75, Nikon Plan Fluor 40/0.75 | |
| GNU Image Manipulation Program (Version 2.10.10) | GIMP | ||
| Illustrator (2017 Creative Cloud) | Adobe | ||
| Image J (Version 1.46r) | NIH | ||
| Microfil | World Precision Instruments | MF 34G-5 | |
| Micromanipulator with universal adaptor and support base | Drummond Scientific | 3-000-024-R | |
| 3-000-025-SB | |||
| 3-000-024-A | |||
| Micropipette Puller | Sutter Instrument | P-30 | |
| Miniprep Kit | Qiagen | 27104 | |
| Motorized z-stage | Applied Scientific Instrumentation | MFC-2000 | |
| Nanoject II injector | Drummond Scientific | 3-000-204 | |
| Powerpoint (Version 15.31) | Microsoft | ||
| Xenopus laevis embryos | Nasco | LM00490 |
References
- Gibson, D. A., Ma, L. Developmental regulation of axon branching in the vertebrate nervous system. Development. 138 (2), 183-195 (2011).
- Alsina, B., Vu, T., Cohen-Cory, S. Visualizing synapse formation in arborizing optic axons in vivo: dynamics and modulation by BDNF. Nature Neuroscience. 4 (11), 1093-1101 (2001).
- Harris, W. A., Holt, C. E., Bonhoeffer, F. Retinal axons with and without their somata, growing to and arborizing in the tectum of Xenopus embryos: a time-lapse video study of single fibres in vivo. Development. 101 (1), 123-133 (1987).
- Sakaguchi, D. S., Murphey, R. K. Map formation in the developing Xenopus retinotectal system: an examination of ganglion cell terminal arborizations. Journal of Neuroscience. 5 (12), 3228-3245 (1985).
- Elul, T. M., Kimes, N. E., Kohwi, M., Reichardt, L. F. N-and C-terminal domains of β-catenin, respectively, are required to initiate and shape axon arbors of retinal ganglion cells in vivo. Journal of Neuroscience. 23 (16), 6567-6575 (2003).
- Wiley, A., et al. GSK-3β and α-catenin binding regions of β-catenin exert opposing effects on the terminal ventral optic axonal projection. Developmental Dynamics. 237 (5), 1434-1441 (2008).
- Jin, T., Peng, G., Wu, E., Mendiratta, S., Elul, T. N-terminal and central domains of APC function to regulate branch number, length and angle in developing optic axonal arbors in vivo. Brain research. 1697, 34-44 (2018).
- Marshak, S., Nikolakopoulou, A. M., Dirks, R., Martens, G. J., Cohen-Cory, S. Cell-autonomous TrkB signaling in presynaptic retinal ganglion cells mediates axon arbor growth and synapse maturation during the establishment of retinotectal synaptic connectivity. Journal of Neuroscience. 27 (10), 2444-2456 (2007).
- Holt, C. E., Garlick, N., Cornel, E. Lipofection of cDNAs in the Embryonic Vertebrate Central Nervous System. Neuron. 4 (2), 203-214 (1990).
- Ohnuma, S. I., Mann, F., Boy, S., Perron, M., Harris, W. A. Lipofection strategy for the study of Xenopus retinal development. Methods. 28 (4), 411-419 (2002).
- Joesch, M., Meister, M. A neuronal circuit for colour vision based on rod-cone opponency. Nature. 532 (7598), 236-239 (2016).
- Meyer, M. P., Smith, S. J. Evidence from in vivo imaging that synaptogenesis guides the growth and branching of axonal arbors by two distinct mechanisms. Journal of Neuroscience. 26 (13), 3604-3614 (2006).
- Li, X., Monckton, E. A., Godbout, R. Ectopic expression of transcription factor AP-2δ in developing retina: effect on PSA-NCAM and axon routing. Journal of Neurochemistry. 129 (1), 72-84 (2014).
- Haas, K., Jensen, K., Sin, W. C., Foa, L., Cline, H. T. Targeted electroporation in Xenopus tadpoles in vivo-from single cells to the entire brain. Differentiation. 70 (4-5), 148-154 (2002).
- Falk, J., et al. Electroporation of cDNA/Morpholinos to targeted areas of embryonic CNS in Xenopus. BMC Developmental Biology. 7, 107 (2007).
- Sive, H. L., Grainger, R. M., Harland, R. M. . Early Development of Xenopus laevis: A Laboratory Manual. , (2000).
- Nieuwkoop, P. D., Faber, J. . Normal table of Xenopus laevis (Daudin). , (1956).
- Zahn, N., Levin, M., Adams, D. S. The Zahn drawings: new illustrations of Xenopus embryo and tadpole stages for studies of craniofacial development. Development. 144 (15), 2708-2713 (2017).
- Piper, M., Dwivedy, A., Leung, L., Bradley, R. S., Holt, C. E. NF-protocadherin and TAF1 regulate retinal axon initiation and elongation in vivo. Journal of Neuroscience. 28 (1), 100-105 (2008).
- Dwivedy, A., Gertler, F. B., Miller, J., Holt, C. E., Lebrand, C. Ena/VASP function in retinal axons is required for terminal arborization but not pathway navigation. Development. 134 (11), 2137-2146 (2007).
- Leung, L. C., Harris, W. A., Holt, C. E., Piper, M. NF-Protocadherin Regulates Retinal Ganglion Cell Axon Behaviour in the Developing Visual System. PLOS One. 10 (10), e0141290 (2015).
- Lee, P. C., He, H. Y., Lin, C. Y., Ching, Y. T., Cline, H. T. Computer aided alignment and quantitative 4D structural plasticity analysis of neurons. Neuroinformatics. 11 (2), 249-257 (2013).

