Conjunctival Commensal Isolation and Identification in Mice
Summary
Presented here is a protocol for the isolation and amplification of aerobic and facultative anaerobic mouse conjunctival commensal bacteria using a unique eye swab and culture-based enrichment step with subsequent identification by microbiological based methods and MALDI-TOF mass spectrometry.
Abstract
The ocular surface was once considered immune privileged and abiotic, but recently it appears that there is a small, but persistent commensal presence. Identification and monitoring of bacterial species at the ocular mucosa have been challenging due to their low abundance and limited availability of appropriate methodology for commensal growth and identification. There are two standard approaches: culture based or DNA sequencing methods. The first method is problematic due to the limited recoverable bacteria and the second approach identifies both live and dead bacteria leading to an aberrant representation of the ocular space. We developed a robust and sensitive method for bacterial isolation by building upon standard microbiological culturing techniques. This is a swab-based technique, utilizing an “in-lab” made thin swab that targets the lower conjunctiva, followed by an amplification step for aerobic and facultative anaerobic genera. This protocol has allowed us to isolate and identify conjunctival species such as Corynebacterium spp., Coagulase Negative Staphylococcus spp., Streptococcus spp., etc. The approach is suitable to define commensal diversity in mice under different disease conditions.
Introduction
The aim of this protocol is to enhance specific isolation of viable and rare aerobic and facultative anaerobic microbes from the ocular conjunctiva to characterize the ocular microbiome. Extensive studies have profiled commensal mucosal communities on the skin, gut, respiratory and genital tracts and show that these communities influence the development of the immune system and response1,2,3. Ocular commensal communities have been shown to change during certain disease pathologies, such as Dry eye disease4, Sjogren’s syndrome5 and diabetes6. Yet, the ability to define a typical ocular surface commensal community is hampered by their relatively low abundance compared to the other mucosal sites6,7,8. This prompts a controversy over whether there is a resident ocular microbiome and if it exists, whether it differs from the skin microbiome and consequently, its local effect on the innate immune system development and response. This protocol can help resolve this question.
Generally, approaches to define the ocular commensal niche are based on sequencing and culture-based techniques4,7,9. 16 S rDNA sequencing and BRISK analysis7 show a broader diversity than culture-based techniques, but are unable to differentiate between live and dead microbes. Since the ocular surface is hostile to many microbes due to tear film’s anti-microbial properties4 generating a large array of DNA fragments, DNA based approaches will detect these artifacts which may skew the data toward identification of dead bacteria as resident commensals rather than contaminants. This results in aberrant commensal identification and characterization of the ocular space as being higher in microbe abundance and diversity10. This makes it difficult to define the resident ocular microbiome via DNA based methods. Whereas, standard culture-based techniques are unable to detect commensals because the load is too low11. Our method improves upon standard practices by using a thin swab that can target the conjunctiva, thus avoiding contamination from neighboring skin, as well as the concept that viable organisms can be enriched by brief culture in nutrient dense media with the goal of resuscitating viable but non-culturable, as well as, enriching for rare viable microbes.
The results, relative abundance of ocular commensals per eye swab, characterize the conjunctiva resident microbiome and are important for comparative purposes. Our data shows that there is a difference between skin and conjunctival microbiota, as well as greater diversity with increased age and a sex specific difference in abundance. Furthermore, this approach has reproducibly found commensal differences in knock-out mice12. This protocol can be applied to describe the ocular microbiome which may vary due to caging practices, geography, or disease state, as well as the local effects of commensal metabolites and products on immune system development and response.
Protocol
All procedures involving mice follow the Institutional Animal Care and Use Committee guidelines. Follow laboratory safety guidelines (as directed by your Institutional Environmental Health and Safety department) when working with microorganisms and potentially contaminated materials. Use appropriate waste receptacles and decontamination procedures prior to disposal of potentially biohazard contaminated materials.
1. Eye swab preparation, work field set-up, mouse eye swabbing and sample enrichment
- Prepare sterile eye swabs
NOTE: Sterile eye swabs are thinly coated wooden toothpicks with a fiber thin layer of cotton batting and made in house.- Autoclave appropriate amount of cotton batting and toothpicks for number of mice to be swabbed.
- Pinch off a half a centimeter-long piece of cotton batting with thumb and forefinger. Tease out batting by pulling on the edges with thumbs and forefingers to form a flat single porous layer, stopping just before the batting falls apart (small bits of cotton dispersed throughout the stretched batting are acceptable).
- Swirl the batting around one of the sharp ends of the toothpick by lightly holding the stretched-out piece on the toothpick tip as it is twisted, see Figure 1. The “completed” eye swab will have a very thin layer of cotton stretched over the tip, extending approximately one half to one centimeter away from the tip.
- Insert “completed” swabs into small beaker, swab side down and cover. Autoclave.
- Set up the workspace
- Prepare anesthesia containing 2 mL of ketamine (100 mg/mL), 400 µL of xylazine (100 mg/ml) and 27.6 mL of sterile saline (0.9% NaCl in dH2O) in a sterile 50 mL sterile centrifuge tube. Filter sterilize and aliquot anesthesia for immediate use (may be stored at 4 °C for 1 month; always allow to equilibrate to room temperature prior to use).
- Clear and clean the work area with disinfectant to minimize contamination.
- Aliquot 0.5 mL of sterile Brain Heart Infusion media (BHI) into labeled 1.5 mL sterile microcentrifuge tubes (1 tube per mouse and control); arrange in microcentrifuge rack. Set the rack on ice.
- Set up work flow as follows (from left to right): mouse anesthetizing station (cage containing experimental mice, empty sterile cage, room temperature anesthesia, 25 G needle and 1 mL syringe), eye swabbing station (aliquoted BHI on ice, sterilized eye swabs, clean paper towels, 70% isopropanol spray) and plating station (room temperature blood agar plates, 10 µL pipette, 10 µL sterile disposable tips, biohazard waste container).
- Eye swabbing
- Administer 10 µL of anesthesia per 1 g of mouse weight dosage of ketamine and xylazine respectively intraperitoneally to each adult mouse and place into an autoclaved cage. Test anesthetization by squeezing hindfoot pad; no movement indicates appropriate anesthetization.
- Assign one hand to only handle anesthetized mice and the other hand to only handle the eye swab and the culture. For swabbing, remove the fully anesthetized mouse from the cage; place on top of clean work surface positioned on its’ side with left eye exposed. Spray gloved hands with isopropanol, dry with clean paper towel.
- Uncap specifically labeled BHI micro-centrifuge tube with dedicated media handling hand. Place the tube back into the micro-centrifuge rack on ice. Dip the cotton coated tip of the eye swab in BHI, withdraw the eye swab from the tube swirling the tip 2 times against the inner tube to remove excess liquid.
- With the mouse handling hand, gently hold the mouse by the scruff of the neck. With the other hand, place the tip of eye swab against the medial conjunctival region of the left eye, see Figure 2A. Lightly depress the eyeball and move the swab in a window washing motion (Figure 2B) back and forth, between the lower eyelid and eye, 10 times. Maintain constant pressure.
- Without touching the fur, gently remove the tip of swab, perpendicular to where it was inserted, and place the swab cotton side down directly into a labeled micro-centrifuge tube containing BHI media. Apply a lubricating eye drop to the swabbed eye.
- Return the mouse to the cage.
- Let the swab stand for 10 to 15 min on ice and remove eye swab with the sterilized gloved hand by mixing the tip in the media for 10 rotations. Withdraw the swab by swirling tip against the inner wall of the micro-centrifuge tube for 5 rotations. Dispose of the swab in biohazard container.
- Repeat steps 1.3.2 to 1.3.7 for each mouse and for control samples (skin or fur swab). Sterilize gloves appropriately between each swab.
- Enrichment
- Enrich the sample by incubating the tube statically for 1 h at 37 °C
- During incubation, label one room temperature Trypticase Soy with 5% sheep’s blood agar plate (TSA plate) per mouse and divide in half.
- After eye swab culture incubation, remove enriched samples from the incubator and place on ice.
- Briefly vortex samples to mix and plate according to the schematic in Figure 3A. Aliquot 10 µL of the sample onto the TSA plate and tilt the plate to form a strip, repeat 2 times.
- On the other side of the plate’s dividing line, dot 10 µL of sample on agar, repeat 9 times.
- Incubate plates at 37 °C for 18 h, 2 days, and 4 days (in a clean chamber that prevents agar plates from drying). Count colonies in strips, note morphology and calculate colony forming units (CFUs) per swab for morphologically similar isolates. Look at dots for unique organisms not captured in strips.
2. Master plate, characterization, and identification of ocular microbes
- Master plate
- Draw grids on the back of a TSA plate (Figure 3B). Pick a colony with a sterile toothpick from each mouse eye swab plate and streak the master plate square (grids), label grid with colony number and mouse information. Pick and plate three different colonies for each morphologically distinct colony.
- Incubate the plate overnight at 37 °C. Continue incubation for 96 hours, checking daily for the appearance of new isolates.
NOTE: The Commensal population will vary based on mouse age, nutrition, disease state and caging practices. Isolate characterization is based on the predominant aerobic and facultative anaerobic bacteria found in our laboratory.
- Characterization
- Duplicate13 plate microbes on Mannitol Salt Agar (MSA) and MacConkey (MAC) plates, incubate overnight at 37 °C. Note growth for each isolate and presence of waxy colonies or yellow colonies with halo on MSA.
- For Gram positive cocci, test with the catalase test13,14. Place a drop of 3% hydrogen peroxide on a clean glass slide. Using a sterile toothpick, pick the corresponding microbe from the eye swab plate. No bubble formation indicates Streptococcus spp., slight bubble formation indicates Corynebacterium spp. and perhaps Aerococcus spp. and rapid bubble formation indicates Staphylococcus spp.
- Identification: MALDI-TOF MS analysis
NOTE: Bacterial identifications are carried out using the MALDI-TOF and the Software Database. This system employs matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for developing spectra profiles which are compared to known bacterial spectra for identification. E.coli, ATCC 8739, is used for system calibration.- Plate bacterial isolates onto TSA plates containing 5% sheep blood and incubate overnight with 5% carbon dioxide at 37 ˚C.
- Using a 1 µL loop, apply a thin layer of the pure bacteria to the MALDI-TOF target slide; 1 µL of MALDI-TOF MS CHCA matrix solution (alpha-cyano-4-hydroxycinnamic acid) is overlaid and allowed to air dry completely before loading the slide into the MALDI-TOF instrument.
- Each isolate is spotted in duplicate.
- Reports containing probability scores greater than 80%, which indicate a high discrimination value, are accepted as reliable results and the bacterial identification accepted.
Representative Results
Representative results for an eye swab plate demonstrating different methods for plating are pictured in Figure 3A showing morphologically diverse isolates from C57BL/6 mouse. For each distinct isolate, the colonies were counted in the strip and the relative abundance, unique Colony Forming Units (CFUs) per eye swab, calculated and plotted for comparison purposes. For microbiological characterization, bacteria were picked from individual mouse eye swab plates to produce a master TSA plate (created by picking morphologically distinct isolates in triplicate from each mouse eye swab plate). From the master plate, on the subsequent day or when growth appears, additional tests were run to characterize or identify the microbes. The master plate was used to provide enough inoculum to expand the respective isolates.
The species were characterized using microbiological techniques and then identified by MALDI-TOF MS analysis. Each isolate from the master plate, was tested by catalase test13,14 and grown on selective media. Since the predominant microbes in our studies are Streptococcus spp., Staphylococcus spp. and Corneybacterium spp., Mannitol Salt Agar (MSA) and Mac Conkey Agar (MAC) were used as the selective agar13. Growth on Mannitol Salt Agar indicates Staphylococcus spp13,16. Since MSA contains phenol red, a yellow halo around the colony indicates mannitol fermentation and classification as Staphylococcus aureus (confirm with alternative tests such as Coagulase test)13. Growth on MacConkey agar indicates Gram negative bacteria, since bile salts and crystal violet are able to transgress the bacterial membrane and inhibit Gram positive bacterial growth13. Waxy growth on MSA indicates the production of mycolic acid and may be due to organisms such as Corneybacterium spp13. Finally, the catalase test differentiates Streptococcus spp. from Staphylococcus spp13,16, and in some cases, a weak positive test may indicate Aerococcus spp15.
To identify isolates, a TSA plate was streaked and incubated overnight in a 5% carbon dioxide and 37 °C environment and MALDI-TOF MS performed. Figure 4 shows the results S. acidominimus (a member of viridans Streptococcus spp.17) and A. viridans. Often Aerococcus spp. are misidentified as the viridans group of streptococci18,19, based on biochemical and phenotypic tests. MALDI-TOF MS was able to identify the isolates with a confidence level of 99.9 with no unidentifiable species.
Sex biased differences can be observed in Figure 5, where significantly different levels of commensal organisms were recovered from male and female C57BL/6 mice. The relative abundance for each isolate was determined. Streptococcus acidominimus, Aerococcus viridans and coagulase negative staphylococcus (CNS) isolate #1 and E. coli spp. were found in all mice, with the males showing higher relative abundance and greater diversity.
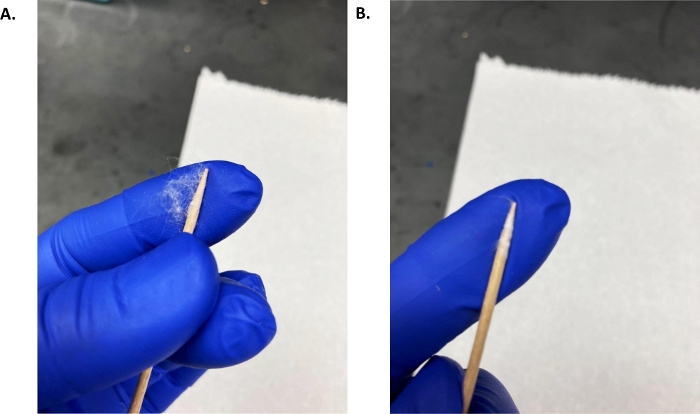
Figure 1: In-house eye swab.
(A) A thin, approximately 1 cm, piece of pulled cotton batting was held between the sterile toothpick point and index finger and the toothpick was rolled while maintaining slight pressure against the index finger until the cotton was completely rolled onto the toothpick tip. (B) Example of an eye swab. Please click here to view a larger version of this figure.
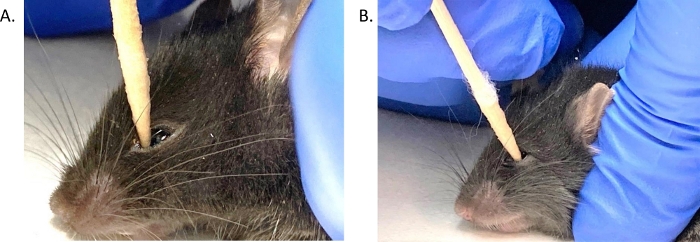
Figure 2: Mouse conjunctival eye swab placement.
(A) The BHI wet eye swab tip was inserted, at a 90˚ angle, into the medial corner of the left eye of an anesthetized mouse, depressing the eyeball. (B) The swab was moved along the conjunctiva while maintaining slight pressure on the eye in a back and forth motion, 10 times. Please click here to view a larger version of this figure.
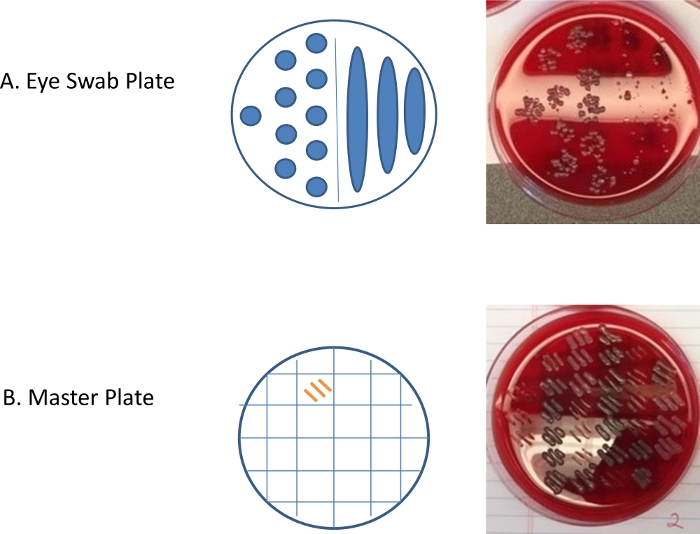
Figure 3: Representative results for the eye swab and master plates.
(A) 10 µL of eye swab inoculated enriched media was aliquoted on the right side of the blood agar plate and tilted 30 to 60 degrees to allow the media to form strips and on the left side of the plate, ten 10 µL dots of the sample was dispensed. The photograph of the incubated plate shows individual colonies and morphological diversity. (B) A master plate of isolates was created by picking a colony from the eye swab plate and streaking within one of the squares (grids). Three morphologically similar colonies are streaked in separate grids. Every grid has three streaks of the same colony that was picked from the mouse eye swab plate. After growth appeared on the plates, the isolate was characterized by selective plating and catalase testing. Please click here to view a larger version of this figure.
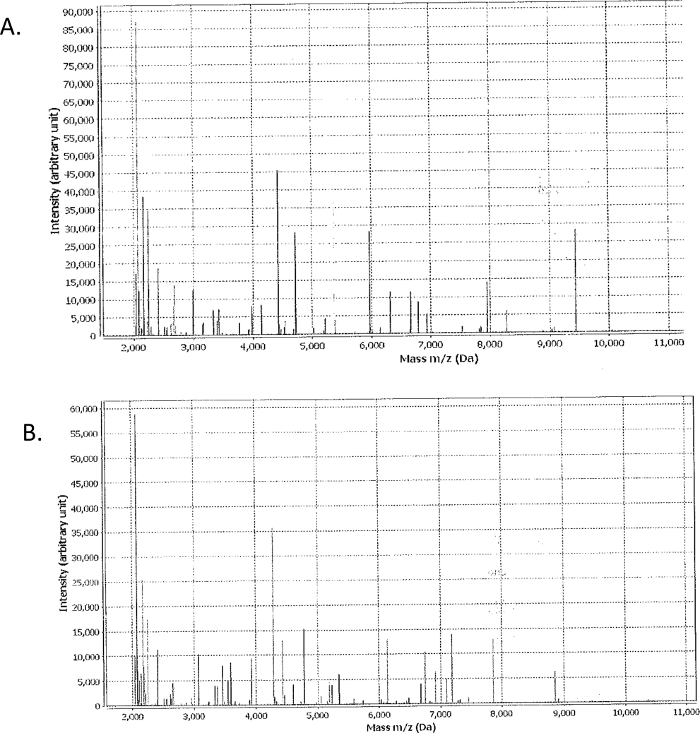
Figure 4: Example of MALDI-TOF MS results.
Each isolate was cultured on TSA overnight, applied to MALDI-TOF MS slide and overlaid with MALDI-TOF MS solution, air dried and spotted in duplicate. Results for Streptococcus acidominimus (A) and Aerococcus viridans (B) were positively identified with a confidence value of 99.9. Please click here to view a larger version of this figure.
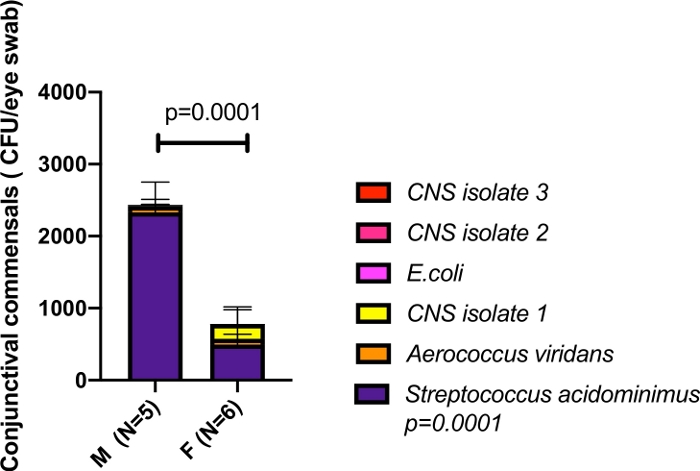
Figure 5: Sex-biased commensal conjunctival growth.
Age-matched C57BL6/N mice were compared for relative commensal diversity. Eyes were swabbed and commensal organisms identified. The most predominant species was Streptococcus acidominimus, which was significantly more abundant in male than female mice (2-way ANOVA, p<0.0001). 5 different CNS isolates were identified, Aerococcus viridans and E.coli. While some of the CNS isolates were not present in all mice, the Streptococcus acidominimus, Aerococcus viridans, CNS isolate 1, and E. coli were found in all mice with distinct abundances. Please click here to view a larger version of this figure.
Discussion
Due to the paucibacterial state of the ocular surface, many laboratories have had difficulty isolating ocular commensals7,20, resulting in low number of samples with growth, low abundance and low diversity8. This method significantly improves upon standard culture practices4,21 by the addition of an enrichment step, as well as a redesigned eye swab and identification by MALDI-TOF MS. The enrichment step addresses low recoverability by amplifying the bacterial load. The significant increase in recoverable bacteria from less than 100 CFUs to the relative abundance of 2,500 CFUs per eye swab for wild type male mice suggests that incubation in nutrient rich media reinvigorates viable but non-cultivatable bacteria22,23, thus permitting the recovery of dormant microbes. Recent culture-based enrichment steps have been incorporated into microbiota molecular sequencing studies to enable discrimination between host and microbial expression signatures24,25,26, and to amplify rare genera. Our protocol enrichment step is the first to be applied to ocular microbiome studies and utilizes enrichment to select live microbes, expand the number of recoverable scarce isolates and perhaps resuscitate viable but non-cultivable commensals. Furthermore, our in house made eye swab allows targeted swabbing of the conjunctiva, reducing contamination from surrounding skin/fur due to its small size and thin pointed tapered shape. The choice of swab coating thickness and material is important21. This laboratory made eye swab is coated with a thin layer of nontoxic cotton batting, which prevents swabbed commensal entrapment within the swab material, as well as, being preferable to calcium alginate eye swabs which can be cytoxic27. Longitudinal tests show that our method is reproducible; with the dominant bacteria recovered from eye swabs including Streptococcus spp., Coagulase Negative Staphylococcus (CNS), and Corynebacterium spp. These results agree with published findings for the dominant facultative anaerobic ocular genera7,9,20 detected via culture based or imaging-based methods. This method has found a broader spectrum of isolates in the conjunctiva, including Escherichia coli and Pseudomonas spp. Furthermore, the identification by MALDI-TOF MS enables better discrimination between genera and species, such as the viridans group of streptococci and Aerococci spp.19,28.
Three steps are critical in maximizing the outcome: eye swab manufacture, eye swab technique and unique isolate selection from the eye swab plate. As discussed above, a very thin flat layer of cotton is important for the initial capture of commensals from the ocular surface as well as for their subsequent release into the enrichment media. Thick or lumpy swabs can lead to the selection of contaminants, as well as retaining isolates within the swab tip. Secondly, continuous depression of the eyeball while swabbing is necessary to minimize contamination from surrounding surfaces. The ideal swabbing technique involves normal (90 degree) insertion of the swab into the medial inferior conjunctival fornix and steady swabbing pressure coupled with slow continuous movement. Finally, the eye swab plate must be monitored daily to select for newly appearing isolates or those growing at a different rate in order to fully capture the representative microbiome.
There are a few protocol limitations that can be addressed with a clear understanding of the results or modification of the protocol. The results are a measure of relative viable commensal abundance, and not actual conjunctival bacterial burden, with the aim of defining the ocular commensal community. The relative abundance and diversity have been used in our studies for comparative purposes in bacterial keratitis, as well as, to investigate the innate immune response in knock-out and wild type mice12. In addition, identification by MALDI-TOF MS depends on a substantial database28, delivering a result of “no ID” for unknown isolate spectra profiles, highlighting the importance of using a current database. Finally, this protocol only detects aerobic and facultative anaerobic bacteria, but its framework is adaptable and can be built upon to help define the ocular microbial space by modification of enrichment media, plate growth conditions and selection medium. By changing the master plate and enrichment growth conditions to anaerobic, then another commonly isolated ocular commensal, Propionibacterium spp. or other anaerobes, may be detected; although, in our hands we saw almost no anaerobic growth.
Perhaps, this framework may be used in conjunction with deep DNA sequencing techniques. A major hurdle in microbe identification via DNA sequencing is the inability to assess viability10. This protocol may provide an orthogonal method by modifying enrichment media and plating conditions to match a candidate isolate’s (identified by DNA sequencing) preferred growth criteria. This may result in a valuable culture based approach to capture commensals from viable but uncultivatable bacteria or present but low abundance species.
Definition of the viable ocular commensal community is essential for examining the ocular commensal expression profile. Broad research suggests that short chain fatty acids and microbial products modulate the ocular immune response3,29,30,31,32,33, and that their action occurs locally. This suggests that not only is distal production, but local production of these factors is important. Also, disease states such as dry eye disease and diabetes change the ocular surface microbiome4,6,33,34. This highlights the need for a method that bridges DNA based sequencing as well as culture-based approaches so the viable ocular microbiome in health and disease can be better defined.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Funding from P30 DK034854 supported VY, LB and studies in the Massachusetts Host-Microbiome Center and funding from NIH/NEI R01 EY022054 supported MG.
Materials
| 0.1 to 10 µl pipet tip | USA Scientific | 1110-300 | autoclave before use |
| 0.5 to 10 µl Eppendorf pipet | Fisher Scientific | 13-690-026 | |
| 1 ml syringe | Fisher Scientific | BD309623 | 1 syringe for each eye swab group |
| 1.5 ml Eppendorf tubes | USA Scientific | 1615-5500 | autoclave before use |
| 1000 µ ml pipet tip | USA Scientific | 1111-2021 | autoclave before use |
| 200 to 1000µl Gilson pipetman (P1000) | Fisher Scientific | F123602G | |
| 25 G needle | Fisher Scientific | 14-826AA | 1 needle per eye swab group |
| 3 % Hydrogen Peroxide | Fisher Scientific | S25359 | |
| 37 ° C Incubator | Lab equipment | ||
| 70 % Isopropanol | Fisher Scientific | PX1840-4 | |
| Ana-Sed Injection (Xylazine 100 mg/ml) | Santa Cruz Animal Health | SC-362949Rx | |
| BD BBL Gram Stain kit | Fisher Scientific | B12539 | |
| Bunsen Burner | Lab equipment | ||
| Clean paper towels | Lab equipment | ||
| Cotton Batting/Sterile rolled cotton | CVS | ||
| Disposable 1 ml Pipets | Fisher Scientific | 13-711-9AM | for Gram stain and catalase tests |
| E.coli | ATTC | ATCC 8739 | |
| Glass slides | Fisher Scientific | 12-550-A3 | for Gram stain and catalase tests |
| Ketamine (100mg/ml) | Henry Schein | 9950001 | |
| Mac Conkey Agar Plates | Fisher Scientific | 4321270 | store at 4 °C until ready to use |
| Mannitol Salt Agar | Carolina Biological Supply | 784641 | Prepare plates according to mfr's instructions, store at 4 °C for 1 week |
| Mice | Jackson Labs | C57/BL6J | |
| Petri Dishes | Fisher Scientific | 08-757-12 | for Mannitol Salt agar plates |
| RPI Brain Heart Infusion Media | Fisher Scientific | 50-488525 | prepare according to directions and autoclave |
| SteriFlip (0.22 µm pore size polyester sulfone) | EMD/Millipore, Fisher Scientifc | SCGP00525 | to sterilize anesthesia |
| Sterile Corning Centrifuge Tube | Fisher Scientific | 430829 | anesthesia preparation |
| Sterile mouse cage | Lab equipment | ||
| Tooth picks (round bamboo) | Kitchen Essentials | autoclave before use and swab preparation | |
| Trypticase Soy Agar II with 5% Sheep's Blood Plates | Fisher Scientific | 4321261 | store at 4 °C until ready to use |
| Vitek target slide | BioMerieux Inc. Durham,NC | ||
| Vitek-MS | BioMerieux Inc. Durham,NC | ||
| Vitek-MS CHCA matrix solution | BioMerieux Inc. Durham, NC | 411071 | |
| Single use eye drops | CVS Pharmacy | Bausch and Lomb Soothe Lubricant Eye Drops, 28 vials, 0.02 fl oz. each |
References
- Arpaia, N., et al. Metabolites produced by commensal bacteria promote peripheral regulatory T cell generation. Nature. 504 (7480), 451-455 (2013).
- Hooper, L. V., Littman, D. R., Macpherson, A. J. Interactions between the microbiota and the immune system. Science. 336 (6086), 1268-1273 (2010).
- Nagpal, R., et al. Human-origin probiotic cocktail increases short chain fatty acid production via modulation of mice and human gut microbiome. Scientific Reports. 8 (1), 12649 (2018).
- Graham, J. E., et al. Ocular pathogen or commensal: a PCR based study of surface bacterial flora in normal and dry eyes. Investigative Ophthalmology and Visual Science. 48 (12), 5616-5623 (2007).
- Wang, C., et al. Sjögren-Like Lacrimal Keratoconjunctivitis in Germ-Free Mice. International Journal of Molecular Sciences. 19 (2), 565-584 (2018).
- Ham, B., Hwang, H. B., Jung, S. H., Chang, S., Kang, K. D., Kwon, M. J. Distribution and diversity of ocular microbial communities in diabetic patients compared with healthy subjects. Current Eye Research. 43 (3), 314-324 (2018).
- Doan, T., et al. Paucibacterial microbiome and resident DNA virome of the healthy conjunctiva. Investigative Ophthalmology and Visual Science. 57 (13), 5116-5126 (2016).
- Kugadas, A., Gadjeva, M. Impact of microbiome on ocular health. Ocular Surface. 14 (3), 342-349 (2016).
- Dong, Q., et al. Diversity of bacteria at healthy human conjunctiva. Investigative Ophthalmology and Visual Science. 53 (8), 5408-5413 (2011).
- Zegans, M. E., Van Gelder, R. N. Considerations in understanding the ocular surface microbiome. American Journal of Opthalmology. 158 (3), 420-422 (2014).
- Fleiszig, S. M., Efron, N. Microbial flora in eyes of current and former contact lens wearers. Journal of Clinical Microbiology. 30 (5), 1156-1161 (1992).
- Lu, X., et al. Neutrophil L-Plastin Controls Ocular Paucibacteriality and Susceptibility to Keratitis. Frontiers in Immunology. 11, 547 (2020).
- Johnson, T. R., Case, C. L. . Laboratory Experiments in Microbiology. , (2010).
- Reiner, K. Catalase Test Protocol. American Society for Microbiology. , (2010).
- UK SMI. . Standards for Microbiology Investigation. UK SMI. , (2014).
- Sharp, S. E., Searcy, C. Comparison of mannitol salt agar and blood agar plates for identification and susceptibility testing of Staphylococcus aureus in specimens from cystic fibrosis patients. Journal of Clinical Microbiology. 44 (12), 4545-4546 (2006).
- Siegman-Igra, Y., Azmon, Y., Schwartz, D. Milleri group streptococcus–a stepchild in the viridans family. European Journal of Clinical Microbiology and Infectious Diseases. 31 (9), 2453-2459 (2012).
- Mohan, B., Zaman, K., Anand, N., Taneja, N. Aerococcus Viridans: A Rare Pathogen Causing Urinary Tract Infection. Journal of Clinical and Diagnostic Research. 11 (1), 1-3 (2017).
- Senneby, E., Nilson, B., Petersson, A. C., Rasmussen, M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry is a sensitive and specific method for identification of aerococci. Journal of Clinical Microbiology. 51 (4), 1303-1304 (2013).
- Wan, S. J., et al. IL-1R and MyD88 contribute to the absence of a bacterial microbiome on the healthy murine cornea. Frontiers in Microbiology. 9, 1117 (2018).
- Ozkan, J., et al. Temporal Stability and Composition of the Ocular Surface Microbiome. Scientific Reports. 7 (1), 9880 (2017).
- Oliver, J. M. The viable but non-culturable state in bacteria. Journal of Microbiology. 43 (1), 93-100 (2005).
- Epstein, S. S. The phenomenon of microbial uncultivability. Current Opinion in Microbiology. 16 (5), 636-642 (2013).
- Whelan, F. J., et al. Culture-enriched metagenomic sequencing enables in-depth profiling of the cystic fibrosis lung microbiota. Nature Microbiology. 5 (2), 379-390 (2020).
- Raymond, F., et al. Culture-enriched human gut microbiomes reveal core and accessory resistance genes. Microbiome. 7, 56 (2019).
- Peto, L., et al. Selective culture enrichment and sequencing of feces to enhance detection of antimicrobial resistance genes in third-generation cephalosporin resistant Enterobacteriaceae. PLoS One. 14 (11), 0222831 (2019).
- Lauer, B. A., Masters, H. B. Toxic effect of calcium alginate swabs on Neisseria gonorrhoeae. Journal of Clinical Microbiology. 26 (1), 54-56 (1988).
- Dubois, D., et al. Performances of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for rapid identification of bacteria in routine clinical microbiology. Journal of Clinical Microbiology. 50 (8), 2568-2576 (2012).
- Kawakita, T., et al. double-blind study of the safety and Efficacy of 1%D-3-Hydroxybutyrate eye drops for Dry Eye Disease. Scientific Reports. 6, 20855 (2016).
- Lee, H. S., Hattori, T., Stevenson, W., Cahuhan, S. K., Dana, R. Expression of toll-like receptor 4 contributes to corneal inflammation in experimental dry eye disease. Investigative Ophthalmology and Visual Science. 53 (9), 5632-5640 (2012).
- Simmons, K. T., Xiao, Y., Pflugfelder, S. C., de Paiva, C. S. Inflammatory response to lipopolysaccharide on the ocular surface in a murine dry eye model. Investigative Ophthalmology and Visual Science. 57 (6), 2444-2450 (2016).
- Miller, D., Ioviano, A. The role of microbial flora on the ocular surface. Current Opinion in Allergy and Immunology. 9 (5), 466-470 (2009).
- Nayyar, A., Gindina, S., Barron, A., Hu, Y., Danias, J. Do epigenetic changes caused by commensal microbiota contribute to development of ocular disease? A review of evidence. Human Genomics. 14 (1), 11 (2020).
- Stevenson, W., et al. Dry eye disease: an immune-mediated ocular surface disorder. Archives of Ophthalmology. 130 (1), 90-100 (2012).

