Experimental Infection with Listeria monocytogenes as a Model for Studying Host Interferon-γ Responses
Summary
This protocol describes how to inoculate C57BL/6J mice with the EGD strain of Listeria monocytogenes (L. monocytogenes) and to measure interferon-γ (IFN-γ) responses by natural killer (NK) cells, natural killer T (NKT) cells, and adaptive T lymphocytes post-infection. This protocol also describes how to conduct survival studies in mice after infection with a modified LD50 dose of the pathogen.
Abstract
L. monocytogenes is a gram-positive bacterium that is a cause of food borne disease in humans. Experimental infection of mice with this pathogen has been highly informative on the role of innate and adaptive immune cells and specific cytokines in host immunity against intracellular pathogens. Production of IFN-γ by innate cells during sublethal infection with L. monocytogenes is important for activating macrophages and early control of the pathogen1-3. In addition, IFN-γ production by adaptive memory lymphocytes is important for priming the activation of innate cells upon reinfection4. The L. monocytogenes infection model thus serves as a great tool for investigating whether new therapies that are designed to increase IFN-γ production have an impact on IFN-γ responses in vivo and have productive biological effects such as increasing bacterial clearance or improving mouse survival from infection. Described here is a basic protocol for how to conduct intraperitoneal infections of C57BL/6J mice with the EGD strain of L. monocytogenes and to measure IFN-γ production by NK cells, NKT cells, and adaptive lymphocytes by flow cytometry. In addition, procedures are described to: (1) grow and prepare the bacteria for inoculation, (2) measure bacterial load in the spleen and liver, and (3) measure animal survival to endpoints. Representative data are also provided to illustrate how this infection model can be used to test the effect of specific agents on IFN-γ responses to L. monocytogenes and survival of mice from this infection.
Introduction
IFN-γ is a cytokine that is crucial for mediating immunity against intracellular pathogens and for controlling tumor growth5. The importance of this cytokine in bacterial resistance is evident in the observation that humans with mutations in the IFN-γ signaling pathway are highly susceptible to infection with mycobacteria and salmonellae6. Similarly, mice deficient in either IFN-γ or the IFN-γ receptor exhibit defects in resistance to mycobacteria7-9 and other intracellular pathogens including L. monocytogenes10,11, Leishmania major12, Salmonella typhimurium13, and certain viruses11. In addition to combatting pathogens, IFN-γ plays a crucial role in host-defense against tumors14. Though higher production of IFN-γ is beneficial in the context of infection or cancer, prolonged production of this cytokine has been linked to the development of systemic autoimmunity15-17 and the acceleration of type I diabetes in the non-obese diabetic mouse model18.
The major sources of IFN-γ include NK cells, NKT cells, γδ T cells, T helper 1 (Th1) cells, and cytotoxic T lymphocytes (CTL)5,19,20. IFN-γ enhances both innate and adaptive immunity by: (1) up-regulating major histocompatibility complex (MHC) class I and II expression, (2) increasing the expression of co-stimulatory molecules on antigen presenting cells, (3) enhancing macrophage phagocytosis and the production of pro-inflammatory cytokines and microbicidal factors (e.g., nitric oxide and reactive oxygen species), (4) promoting the differentiation of naïve CD4+ T cells into Th1 effector cells, (5) promoting antibody class switching to immunoglobulin (Ig)2a and IgG3 (in mouse), (6) inducing the production of chemokines to recruit immune cells to sites of infection, and (7) enhancing NK cell and CTL responses5,19. Given the crucial importance of IFN-γ in the host response to pathogens and tumors, recombinant IFN-γ has been tested as a treatment for various infections and malignancies (reviewed in19). However, because systemic administration of IFN-γ or the Th1 promoting cytokine interleukin-12 (IL-12) is associated with side effects and dose-related toxicity19,21, there is interest in developing alternative strategies to increase IFN-γ production by immune cells. Development of new biologics and small molecules requires in vivo screening tools to test whether such agents increase IFN-γ production during an immune response and whether this translates into meaningful biological effects such as increases in animal survival.
Experimental infection of mice with the gram-positive bacterium L. monocytogenes has been an instrumental model for deciphering the role of IFN-γ in host-immunity against intracellular pathogens1,22. Infection of mice with the pathogen intravenously or intraperitoneally (i.p.) leads to the rapid dissemination of the bacteria to the spleen and liver, where they become internalized by resident macrophages and hepatocytes with peak bacterial loads in the spleen occurring between 3 and 4 days post-infection1,3,22. Production of IFN-γ by NK cells is important for macrophage activation and early resistance against the pathogen3; however at high infectious doses, production of IFN-γ can also be detrimental to pathogen clearance23. NKT cells are also a source of IFN-γ in the spleen and liver during early control of pathogens2,24 and this production has been shown to amplify IFN-γ production by other cell types including NK cells2. On the other hand, later-acting adaptive T lymphocytes, CD8+ T cells in particular, are important for mediating the clearance of the pathogen and providing protection against re-infection1,4,22.
This infection model has been attractive to researchers for a number of reasons (reviewed in1). First, infection with the pathogen is highly reproducible and induces a strong Th1 and cellular immune response. Secondly, during sublethal infection, bacterial load is concentrated in the liver and spleen where it can be easily measured. Thirdly, the pathogen can be safely handled under Biosafety Level 2 (BSL2) conditions. Fourthly, the organism and the immune response that it generates have been extensively characterized. Finally, a variety of mutant and genetically-modified strains have been developed that are available for use.
Described here is a basic protocol for inoculation of C57BL/6J mice with the EGD strain of L. monocytogenes25 and for measuring IFN–γ responses by NK, NKT, and adaptive lymphocytes post-infection. Also described is how to measure bacterial load in the spleen and liver after sublethal infection and to carry out survival studies after infection with a modified LD50 dose of the pathogen. Finally, representative data are shown of how this protocol can be used to screen the effect of new treatments on IFN-γ responses and mouse survival from L. monocytogenes infection.
Protocol
Safety Statement
This protocol describes infection of mice with live L. monocytogenes. The pathogen is handled safely under BSL2 conditions by trained personnel who are not immunocompromised. Immunocompromised people include pregnant women, the elderly and individuals who are HIV-infected or have chronic conditions that require treatment with immunosuppressive therapy. Personnel should don a protective lab coat or gown, gloves, mask, and eye protection while handling infected samples. The work described herein was performed under BSL2 conditions under a certificate (#32876) that was issued by the University Health Network (UHN) Biosafety office. Carcasses from infected mice or any unused tissues were double-bagged and disposed of in biohazard waste. Cages from infected mice were also decontaminated by autoclaving.
Ethics Statement
Mice were maintained and infected in a quarantine room within UHN animal facilities and were cared for in accordance with the guidelines set by the Canadian Council on Animal Care. All procedures on mice were carried out under animal use protocol #3214 that was approved by the UHN animal care committee. Due to ethical considerations, death was not used as an endpoint for survival studies. The modified LD50 dose reported here for L. monocytogenes infection was determined to be the dose at which 50% of the mice reached specific endpoints, which consisted of a 20% loss in body weight or showing at least two of the following clinical signs: lethargy, ruffled fur, hunched posture, labored breathing, dull or sunken eyes. Mice were euthanized when they reached endpoints via exposure to carbon dioxide (CO2) according to UHN facility guidelines.
1. Preparation of Glycerol Stocks for Long-term Storage
NOTE: This procedure describes how glycerol stocks of the EGD strain of L. monocytogenes are prepared from an original glycerol stock. Steps that have the potential to generate aerosols should be performed within a certified biosafety cabinet (BSC).
- Prepare brain heart infusion (BHI) agar plates for bacterial growth. For this add 3.8% (w/v) BHI broth and 1.5% (w/v) agar to double distilled H2O (ddH2O). Autoclave liquid. Once the agar cools to 50 °C, dispense liquid into bacterial petri dishes (25 ml/dish) and let plates dry (uncovered) in the BSC for 1 hr.
NOTE: Transfer BHI agar into a 50 °C water bath after autoclaving to avoid solidification prior to pouring plates. Store BHI plates at 4 °C upside down (with media side on top) until ready for use. - Prepare liquid BHI media. For this, mix 3.8% (w/v) BHI broth in ddH2O. Autoclave.
- Remove frozen glycerol stock of the L. monocytogenes EGD strain from the -80 °C freezer and thaw to room temperature.
- Dip a sterile pipette tip in the thawed glycerol stock and immediately streak the tip back and forth across a section of a BHI plate. This is the primary streak.
- Turn the plate by 90 °C and using a fresh pipette tip, drag through the first streak and spread it to the next ¼ of the plate (this is the secondary streak). Repeat once more to make the tertiary streak.
- Turn plate upside down and incubate at 37 °C overnight. Single uniform colonies should be obtained in the last set of streaks and visible between 16 and 24 hr.
- Dispense 10 ml of sterile BHI broth into a sterile vented 50 ml tube. Pick one colony of L. monocytogenes from the plate using a sterile pipette tip and inoculate the broth. Incubate the culture in a 37 °C orbital shaking incubator overnight or until OD600 = 1.0 with settings at 225 rotations per min (rpm).
NOTE: Glass or disposable plastic Erlenmeyer flasks can also be used to culture bacteria. Regardless of the type of container used, make sure that it is sterile, vented and that the volume of culture does not exceed 20% of the total volume of the container to ensure appropriate aeration of the bacteria. - Prepare glycerol stocks by mixing sterile 100% glycerol with overnight bacterial liquid culture at a 1:1 ratio. Distribute the bacterial/glycerol mixture into 2 ml cryogenic vials (500 μl/vial) and transfer vials to -80 °C freezer for storage.
NOTE: Bead stock methods can also be used in place of glycerol stocks to store bacteria. By this method, porous microbeads are inoculated with a pure culture of L. monocytogenes and are stored at -80 °C. Each bead can be used to inoculate a fresh culture as needed. See materials list for further information.
2. Determination of Growth Curve of L. monocytogenes in Day Culture
NOTE: This procedure describes how to generate the growth curve for L. monocytogenes that is used to estimate the colony forming units (CFU) for infection studies. All steps that have the potential to generate aerosols should be performed within a certified BSC.
- Take 100 µl of overnight culture generated in Step 1.7 to 10 ml BHI media in a vented 50 ml tube and grow at 37 °C in a shaking incubator (225 rpm, tilted at 45° angle). Use a non-inoculated tube as a control.
- Take 0.5 ml samples of the culture at hourly intervals (1, 2, 3, 4, 5, 6 hr, etc.). Dilute each aliquot 1:1 (v/v) with BHI media in a plastic cuvette. Pipette up and down to mix. Measure the optical density (OD) at 600 nm (OD600) using a spectrometer. Continue culturing bacteria until OD600=1.
- At the same time, take a 100 μl sample of the culture and dilute with 900 μl BHI media in a sterile 1.5 ml microcentrifuge tube (this is the 10-1 dilution). Centrifuge the bacteria at 6,000 x g for 5 min, and aspirate the supernatant.
- Wash bacteria twice by resuspending the pellet in 1 ml of BHI media, centrifuging for 5 min at 6,000 x g, and then aspirating the supernatant. Resuspend the pellet in 1 ml BHI media. Prepare a 10-fold dilution series of this sample in BHI media (10-2 to 10-9). Spread 100 μl of each diluent onto separate BHI agar plates. Incubate plates overnight at 37 °C in an incubator.
- The next day, pick plates that have between 30 – 300 colonies. Discard the rest. Count the colonies on these plates. Table 1 shows an example of counts obtained in an aliquot that was taken when OD600 = 0.84. In this example, one of the plates (i.e., 10-6 dilution) had colony counts between 30 – 300 and was used for the CFU/ml calculation.
NOTE: Plates with greater than 300 colonies are not used, since overcrowding can hinder bacterial growth and also makes it difficult to discern and enumerate individual colonies. Plates with counts < 30 are also not used because small errors in dilution technique or the presence of contaminants can have a large impact on the precision of counts at the lower end of the range. - Divide the number of colonies by the volume plated and then multiply by the dilution factor to obtain the CFU/ml value for a particular dilution. In the example in Table 1, the count at the 10-6 dilution was 70. Divide this value by 0.1 ml to get the CFU/ml value for the diluted culture. Then multiply this value by the dilution factor (106) to obtain the CFU/ml value of the undiluted culture (7.0 x 108).
- Plot the OD600 (y-axis) versus time in h (x-axis) to identify the logarithmic phase of growth26.
NOTE: This growth curve provides an estimate of the CFU/ml of the day culture when grown to a certain OD reading. Choose a OD600 reading that is in the logarithmic phase of growth that can be used as a target OD600 for growing day cultures. These data now can be used to estimate the CFU in a culture for preparation of inoculum (Procedure 3).
3. Preparation of the Inoculum for Experimental Infection with L. monocytogenes
NOTE: This procedure describes the preparation of the infectious inoculum from a day culture that was started from an overnight culture (prepared in Procedure 2). All of these steps are performed in the BSC unless otherwise indicated.
- Calculate the number of CFU required for infection based on the number of mice and experimental design of the study. Add an appropriate volume of BHI media to a sterile vented Erlenmeyer flask or culture tube.
NOTE: The CFU of bacteria prepared will be dependent on the type of experiment performed. For studying NK and NKT cell responses during infection, each mouse is inoculated with 105 CFU of bacteria (Procedure 6). If studying adaptive T cell responses to infection or measuring bacterial load, each mouse is inoculated with 2 x 104 CFU of bacteria (Procedure 8). If studying survival to endpoints, each mouse is inoculated with the LD50 dose of the pathogen (which is 105 CFU for males and 1.5 x 105 CFU for females, see Procedure 9). - Inoculate the tube containing BHI media with 100 μl of overnight culture. Incubate the culture in a 37 °C orbital shaking incubator (225 rpm) until target OD600 is reached. Transfer culture contents into a sterile centrifuge tube.
- Centrifuge bacteria into a pellet for 5 min at 6,000 x g using a centrifuge. Aspirate the supernatant using a vacuum attached to a trap flask containing bleach.
- Wash pellet twice with 1x phosphate buffered saline (PBS), centrifuging (5 min at 6,000 x g) in between.
- Aspirate the second wash and dilute bacteria at the appropriate concentration in 1x PBS to deliver the CFU of interest to each mouse in a 200 µl volume.
NOTE: It is best to use a commercial source of sterile 1x PBS for washing bacteria and for preparation of the inoculum, since lab glassware can introduce immunological contaminants such as lipopolysaccharide.
4. Experimental Infection of Mice with L. monocytogenes
NOTE: This procedure describes how to infect mice with the inoculum prepared in Procedure 3 and how to verify the CFU delivered in the inoculum. Handling of mice and injections are performed in a BSC.
- Order a sufficient number of male or female C57BL/6J mice for your experiment. Also order mice to serve as uninfected controls.
- Allow mice to acclimatize for 1 week prior to bacterial inoculation.
NOTE: This is because the stress associated with transport of the animals can trigger a transient increase in stress hormone production and lymphopenia27,28. - On the day of inoculation, obtain a baseline body weight for each mouse and record it in the lab notebook.
- In the BSC, mix the bacterial suspension up and down using a sterile pipette to ensure that the bacteria are evenly distributed and then take up 200 µl of the inoculum into a 1 ml safety engineered syringe fitted with a 25 G needle.
- Inject a mouse i.p. with 200 μl of prepared inoculum (e.g., 105 CFU for NK cell infection, Procedure 6). For this procedure, scruff mice with the less dominant hand by grabbing the loose skin around the mouse's shoulders. After ensuring that the mouse is well-restrained, inject the mouse in the lower quadrant of the abdomen, just lateral to the midline to avoid the bladder.
- Dispose of the needle and syringe in a biohazard sharps container.
- Repeat steps 4.3 – 4.6 until all mice are injected. Conduct similar steps with 1x PBS injected mice (non-infected controls).
NOTE: Since the CFU is an estimate based on the growth curve, it is also good practice to check the actual CFU in the inoculum. For this, prepare 3 – 4 different dilutions of the prepared inoculum (using 10-fold dilution series) that you expect will result in countable colonies. Spread 100 μl of each diluent onto a BHI agar plate and incubate overnight at 37 °C. Count the colonies and calculate the actual CFU/ml as described in Procedure 2.
5. Preparing Heat-killed L. monocytogenes for Immune Studies
NOTE: All steps that have the potential to generate aerosols are performed within the BSC.
- Grow day culture until OD600 values are reached that are within the logarithmic phase. Dispense culture into 1.5 ml microcentrifuge tubes.
- Incubate tubes in a 70 °C in a water bath for 1 hr to kill bacteria.
- Wash bacteria twice with 1x PBS as in Steps 3.3 and 3.4. Resuspend in sterile complete RPMI media containing fetal calf serum (FCS) (see Supplemental File 1 for recipe) at a concentration of 4 x 106/ml. Aliquot killed bacteria into 2 ml sterile cryogenic vials and store at -80 °C.
- Confirm the death of the bacteria by spreading 100 μl of heat-killed bacteria preparation onto BHI agar plates and incubating overnight at 37 °C.
NOTE: These heat-killed bacteria should be ready for stimulating lymphocytes in culture in Procedure 8. If there are any colonies growing on the BHI agar plate, repeat heat-killing procedure.
6. Measurement of IFN-γ Responses by NK and NKT Cells during Infection
NOTE: This procedure describes how to measure the IFN-γ responses by NK and NKT cells in mice at 24 hr after infection with 105 CFU of the L. monocytogenes. This dose is used because it induces robust IFN-γ responses by NK and NKT cells in the spleen24. Conduct all steps in the BSC. To help maintain cell viability, keep cells on ice whenever possible and use ice-cold buffers.
- Inoculate mice as described in Procedure 4 with 105 CFU of the L. monocytogenes. At the same time, inject non-infected control mice i.p. with an equal volume of 1x PBS.
- Euthanize mice at 24 hr post-inoculation by CO2 inhalation according to institutional guidelines.
- Lie each mouse on its right side and wet down the skin with 70% ethanol using a squeeze bottle.
- Using aseptic or sterile forceps and tough-cut scissors, incise the skin just below the bottom of the rib cage.
- Spray down the exposed muscle layer with 70% ethanol. The spleen should be visible underneath the muscle layer (open arrow head in Figure 1).
- Using aseptic or sterile forceps and fine scissors, incise the muscle layer to reveal the spleen. Gently grab the spleen with the forceps and use fine scissors to cut the spleen away from surrounding connective tissue.
- Place the spleen in a 15 ml conical tube containing sterile 1x PBS.
- Half-fill sterile petri dishes with sterile 1x PBS. Dissociate the spleen through a 70 μm nylon cell strainer into the petri dish using the flat end of a sterile 3 ml syringe.
- Transfer the splenocyte suspension into a clean sterile 15 ml conical tube using a sterile 10 ml serological pipette.
- Centrifuge samples at 335 x g for 10 min at 4 °C.
- Aspirate the supernatant into a trap flask containing bleach. Loosen the cell pellet by flicking the tube with a finger or by dragging the bottom tube back and forth along a corrugated surface (e.g., air flow vent in the BSC).
- Lyse red blood cells by adding 1.5 ml of Ammonium-Chloride-Potassium (ACK) lysis buffer (see Supplemental File 1 for recipe) to each spleen. After exactly 1 min and 15 seconds, fill the tube with 1x PBS to stop the cell lysis.
- Centrifuge cells as described in step 6.10. Aspirate the supernatant and resuspend the cell pellet in 10 ml of fluorescence-activated cell sorting (FACS) Buffer (sterile 1x PBS containing 2% FCS).
- Count the cells using a hemocytometer. For this, take two aliquots of cell suspension for counting. Add 15 µl of each cell suspension to an equal volume of Trypan blue (0.04%, made by diluting 0.4% trypan blue solution in ddH2O). Load 15 µl of each cell/Trypan blue suspension into the chamber of the hemocytometer (i.e., one in the top chamber, one in the bottom chamber).
- Using a microscope, count all non-blue cells in five large squares of the central grid in each chamber (Figure 2). Take this count and divide it by 10 to obtain the number of cells in 106/ml. In the example in Figure 2, the count is 215 cells in the 5 squares; therefore, the cell concentration is 21.5 x 106/ml. Average the cell concentrations obtained from the two samples.
- Seed 1 x 106 cells per well/stain in a 96-well round-bottom plate for flow cytometry staining. Make sure to also seed cells for unstained and fluorescence minus one (FMO) controls. See recommended flow cytometry staining panel in Table 2.
NOTE: For the following steps, it is recommended to keep cells on ice or at 4 °C and to protect cells from light with foil when fluorochromes are present. A multichannel pipette can be used to dispense liquids into 96-well staining plates to speed up processing. Be careful not to disturb the cell pellet when aspirating the supernatant from the centrifuged plate. Staining can also be done in FACS tubes if the centrifuge is not fitted with plate adapters. All centrifuge steps from this point on are done at 456 x g for 5 min at 4 °C. - Centrifuge the plate and then wash cells twice with FACS buffer. One wash is done by adding 200 μl of FACS buffer to each well, centrifuging the plate, and then aspirating the supernatant.
- Perform blocking step by adding 50 μl/well of FACS buffer containing anti-mouse CD16/CD32 (purified Fc block) (0.5 µg). Incubate cells at 4 °C for 15 min. Wash cells once in 1x PBS as described above.
- Add 100 μl viability dye (fixable viability dye diluted 1:1,000 in 1x PBS) to cells. Stain cells at 4 °C in the dark (in refrigerator) for 30 min. Wash cells twice in FACS buffer as described above.
- After a second wash, add 100 μl of cell surface antibodies or tetramers to respective wells according to the staining panel described in Table 2. Stain cells at 4 °C in the dark (in refrigerator) for 30 min. At this time, also add antibodies for staining single positive and FMO controls.
NOTE: Regarding single positive controls, it is recommended to either use splenocytes that are stained with various fluorochrome versions of the CD4 antibody or commercial compensation beads that are stained with the antibodies used in the panel. Prior to conducting this staining procedure, all FACS antibodies should be titrated in test studies to determine optimal concentrations for staining. - Wash cells twice in FACS buffer as described above and then fix cells by resuspending them in 50 μl of 4% paraformaldehyde (16% paraformaldehyde stock diluted in ddH2O) and incubating for 10 min at room temperature. CAUTION: The paraformaldehyde is toxic and should only be handled in the fume hood.
- Wash cells twice in FACS buffer, centrifuging in between. Resuspend cells in FACS buffer. Continue to next step or store cells in the refrigerator protected from light for up to three days.
- Centrifuge cells, remove the supernatant, and wash cells twice with 150 μl of 1x Permeabilization/Wash Buffer (Perm/Wash buffer), centrifuging in between. The Perm/Wash Buffer is prepared from a 10x stock by diluting 1:9 (v/v) in ddH2O.
- After the second wash, resuspend the cells in 150 μl of Perm/Wash Buffer and incubate for 15 min at 4 °C in the dark.
- Centrifuge cells again and then aspirate the supernatant. Resuspend cells in 50 μl of 1x Perm/Wash buffer containing anti-IFN-γ and incubate for 1 hr at 4 °C in the dark.
- Wash cells twice with 1x Perm/Wash buffer, centrifuging in between. Resuspend cells in 250 µl of FACS buffer and then transfer cells to FACS tubes.
- Proceed to flow cytometry acquisition using a flow cytometer that has an appropriate laser configuration and filter set to discriminate fluorochromes used in the staining panel described in Table 229. Collect at least 200,000 events per sample and 10,000 events for compensation controls.
- Apply compensation matrix and analyze data using flow cytometric analysis software30.
7. Measurement of Bacterial Load in the Spleen and Liver at the Time of Peak Infection
NOTE: All steps are performed within a BSC unless otherwise noted.
- Inoculate mice i.p. with 2 x 104 CFU of the pathogen using procedures described in Procedure 4.
- On day 3 post-infection, prepare sterile 1.5 ml microcentrifuge tubes, each containing 500 μl of ice-cold sterile 0.1% Triton X-100 in 1x PBS and 0.2 – 0.3 g of 1.5 – 2 mm acid-washed sterile glass beads. Weigh each tube.
NOTE: Glass beads are acid washed by incubating in 10% acetic acid in a beaker on a magnetic stirrer for 1 hr. These beads are then extensively washed with ddH2O to remove acid, are air-dried, and then autoclaved prior to use. - Euthanize mice by CO2 exposure.
- For dissection of organs, lay the animal on its back on a dissecting board and pin the limbs of the mouse to the board using 25 G needles. Disinfect skin by wetting with 70% ethanol.
- Using sterile tough cut scissors, make a midline incision in the skin from the groin to the mid chest and then from the mid groin towards each knee and from the mid chest towards each elbow. Blunt dissect and reflect back the skin, pinning it open using 25 G needles.
- Disinfect muscle layer by wetting it with 70% ethanol and then using sterile fine scissors make a midline incision in the peritoneal wall. Grab the xiphoid process with forceps. Then, using the same fine scissors, make cuts in the peritoneal wall from the xiphoid process laterally on each side, following the rib cage, just below the diaphragm to reveal the liver.
- Cut out a ~100 mg piece of the liver (use the same lobe for all mice) using sterile scissors and place it in a pre-weighed 1.5 ml microcentrifuge tube.
- Use forceps to gently push aside the organs on the left side of the peritoneal cavity to visualize the spleen. Gently grab the spleen with a pair of forceps and release it from the peritoneal cavity by cutting away the surrounding connective tissue.
- Place the spleen in the pre-weighed 1.5 ml microcentrifuge tube containing beads. Transport tissues to the laboratory in a leak-proof container containing ice. Re-weigh the tubes containing the organs to determine the tissue weights in mg.
- Homogenize the tissues by shaking the tubes using a bead mill homogenizer for 3 min at frequency of 30 Hertz.
NOTE: The bead mill method is preferred for homogenization as it is amenable for processing a large number of samples and creates less mess and potential exposure to the pathogen. However, automatic homogenizers or autoclaved 2 ml manual glass tissue homogenizers could be used as an alternative. - Prepare a 10-fold dilution series of the homogenates in 0.1% Triton-X-100 in 1x PBS, ranging from (ranging from undiluted to 10-7).
- Spread 100 μl of each diluted homogenate onto a BHI agar plate (in duplicate) using a sterile spreader. Transfer plates to a 37 °C incubator and incubate overnight.
- Keep the plates that contain between 30 and 300 colonies/plate, discard the rest. Count colonies on each plate and determine the mean number of colonies for duplicate spreads.
- Calculate the CFU/mg according the following equation:
CFU/mg = CFU/ml in the homogenate, multiplied by the ml of homogenate prepared, divided by the mg weight of the tissue homogenized.
NOTE: For example, if a mean of 30 colonies were counted after plating 100 μl of 10-2 diluted homogenate prepared from a 120 mg piece of liver that was homogenized in 0.5 ml, the calculations would be as follows:
CFU/ml = 30 colonies x 100 (dilution factor) / 0.1 ml (volume spread) = 30,000 CFU/ml.
CFU/mg = 30,000 CFU/ml x 0.5 ml homogenate / 120 mg tissue = 125 CFU/mg.
8. Effects of L. monocytogenes on IFN-γ Responses by CD4+ and CD8+ Cells
NOTE: This procedure describes how to measure IFN-γ production by splenic CD4+ and CD8+ T effector cells harvested at the time of the peak of the adaptive immune response (~ 7 d post-infection) using two methods: (1) flow cytometry to measure IFN-γ by CD4+ and CD8+ cells by intracellular cytokine staining, and (2) ELISA to measure total IFN-γ levels produced by splenocytes (includes all T cells). Procedures are performed within the BSC.
- Infect mice by injecting i.p. with 2 x 104 CFU of the pathogen using procedures described in Procedure 4.
- On day 7 post-infection, euthanize mice by CO2 inhalation according to institutional guidelines.
- Dissect the spleen (as described above) and place in a 15 ml conical tube containing sterile 1x PBS. Transport the tubes to the laboratory in a leak-proof container containing ice.
- Process the spleens into a single cell suspension, lyse red blood cells as described in section 6, and then resuspend cells in complete RPMI media containing 10% FCS. Count cells using a hemocytometer.
- Set up cultures for measurement of IFN-γ responses. For this dispense cells (4 x 106 in 1 ml/well) into 24-well plates together with and equal number (4 x 106 or 1 ml/well) of thawed heat-killed L. monocytogenes (prepared in Procedure 5). Transfer cells to a 37 °C incubator.
- After 20 hr of incubation, add 0.66 μl/ml of protein transport inhibitor to wells and continue incubations.
- Four hr later, transfer plate to BSC and collect 500 µl of culture supernatant and freeze (at -80 °C) for the later measurement of IFN-γ levels using a commercial enzyme-linked immunosorbent assay (ELISA) kit31. Then collect cells into a sterile 15 ml tube. Wash wells with 1x PBS and pool this wash together with collected cells.
- Conduct cell-surface staining and intracellular staining for IFN-γ on CD4+ and CD8+ cells as described in Procedure 6 except use the staining panel described in Table 3. Proceed to flow cytometer acquisition (collecting 200,000 events/sample) and analyze data29 using flow cytometry analysis software30.
9. Measuring Mouse Survival to Endpoints after L. monocytogenes Infection
NOTE: This procedure describes the effect of an agent on mouse survival to endpoints post-infection with the modified LD50 dose of the pathogen. All these procedures are conducted in the BSC in the animal facility.
- Inject mice i.p. with the modified LD50 dose of L. monocytogenes as described in Procedure 4. This was determined to be 105 CFU (for male) or 1.5 x 105 CFU (for female)31.
- Follow mice twice daily for clinical signs and record these signs and animal body weights in a lab notebook. Euthanize mice if they show a 20% loss in body weight or two clinical signs of listeriosis (lethargy, ruffled fur, hunched posture, labored breathing, dull or sunken eyes).
- After 14 days, euthanize surviving mice via CO2 inhalation.
- Prepare Kaplan-Meier plots of the data by plotting the percent survival of each group against time32.
NOTE: If mice succumb to infection (meet endpoints described in Ethics Statement), this usually occurs by day 5 post-infection (Figure 7).
Representative Results
Figure 3 presents some typical flow cytometry staining of IFN-γ in splenic NK and NKT cells at 24 hr post-infection with 105 CFU of the pathogen. This figure also illustrates the gating strategy for the staining panel described in Table 2. Figure 4 shows some representative data that were obtained in one experiment where male mice were treated with the PPARα antagonist IS001 or vehicle control, infected with 105 CFU L. monocytogenes, and then analyzed for IFN-γ in NK and NKT cells after 24 hr. This figure shows that treatment with IS001 boosted IFN-γ responses by NKT cells, but not NK cells after infection with the pathogen. Figure 5 shows representative staining for IFN-γ in splenic CD4+ and CD8+ T cells at 7 days post-infection after re-stimulation ex vivo with heat-killed pathogen. This figure also shows the gating strategy for the staining panel described in Table 3. Figure 6 shows representative data that were obtained in one experiment where male mice were treated daily with the PPARα antagonist IS001 or vehicle control, infected with a sublethal dose of L. monocytogenes, and analyzed at 7 days post-infection. This experiment shows that treatment with IS001 enhanced IFN-γ responses by both CD4+ and CD8+ lymphocytes. Figure 7 shows representative data from a study that investigated the effect of the PPARα antagonist IS001 on mouse survival to endpoints after infection with the modified LD50 dose of the pathogen. Plotted is the percent survival of mice against time post-infection. This figure shows that treatment with IS001 increased the survival of male mice to endpoints. Together these data illustrate how this model can be applied to investigate the effects of new drugs or treatments on IFN-γ responses in vivo and to explore how these immune changes impact animal survival from infection.
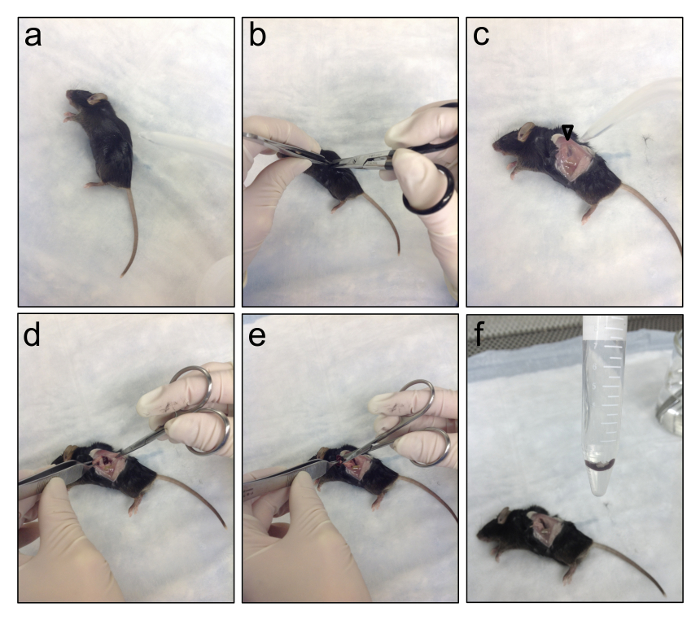
Figure 1. Dissecting the Spleens from Infected Mice. This series of photos shows how to dissect the spleen from a dead mouse. (a) Lie the mouse on its right side and spray down the skin with 70% ethanol. (b) Using aseptic or sterile forceps and tough-cut scissors, incise the skin just below the bottom of the rib cage. (c) Spray down the exposed muscle layer with 70% ethanol. The spleen should be visible underneath the muscle layer (open arrow head). (d) Using aseptic or sterile forceps and fine scissors, incise the muscle layer to reveal the spleen. (e) Gently grab the spleen with the forceps and use fine scissors to cut the spleen away from surrounding the connective tissue. (f) Place the spleen in a 15 ml conical tube containing sterile 1x PBS. Please click here to view a larger version of this figure.
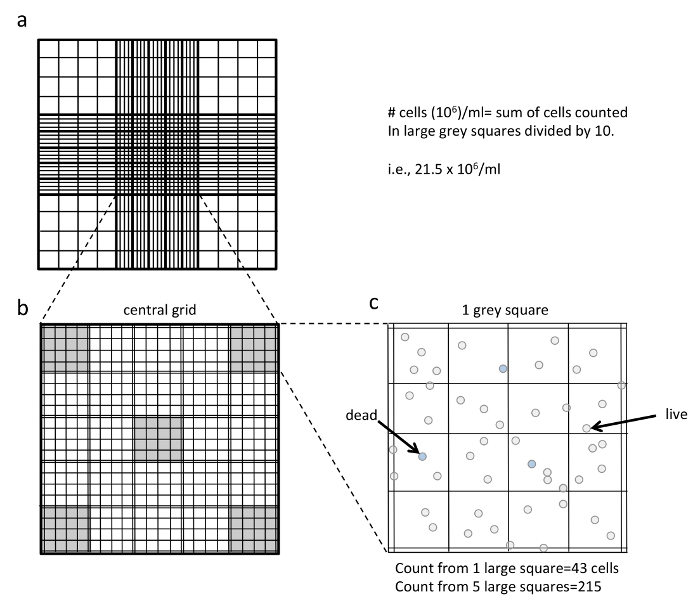
Figure 2. Counting Splenocytes using a Hemocytometer. (a) shows the central grid of the hemocytometer. (b) shows an enlarged view of the central grid that contains 25 large squares (that each contain 16 smaller squares). The five large squares used for counting are highlighted in grey (4 corner squares plus the center square in the central grid). (c) shows an enlarged view of one of the large grey squares. To determine the cell volume in 106/ml, first count all the viable cells within the five large grey squares. In the example shown, this count is 215. When counting, make sure to only count all of the clear (non-blue) cells, including those that are touching the double lines on the right and bottom of the grid. Do not count the cells touching the double lines on the left and top of the grid. Take the total five square count and divide it by 10 to obtain the number of cells in 106/ml. In the example, 215 divided by 10 is 21.5 x 106 cells/ml. Note that these calculations only work if you are counting 5 of the large squares as highlighted and dilute your cells 1:1 in trypan blue. Please click here to view a larger version of this figure.
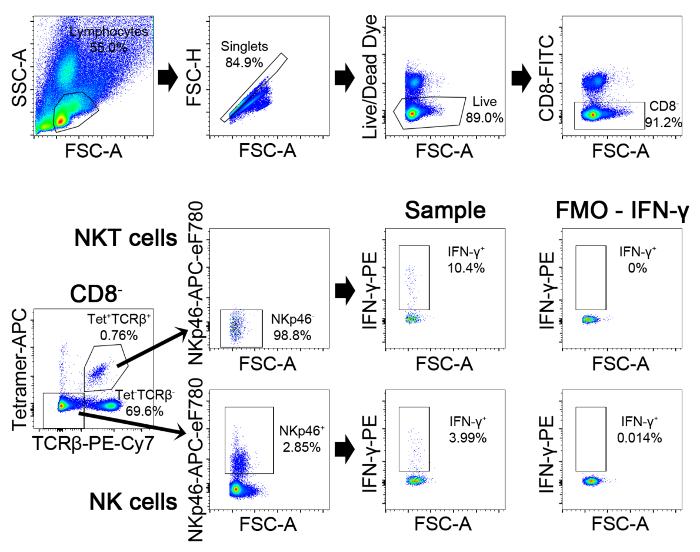
Figure 3. Gating Strategy for Detection of IFN-γ Production in NK and NKT cells. First gate on lymphocytes on FSC-A by SSC-A plot. Then gate on those events that are on the diagonal on the FSC-H/FSC-A plot. These are the singlets. Then gate on live (AmCyan–) and CD8– cells. Then plot the tetramer staining against TCRβ. The NKT cells are within the double positive population and the NK cells are within the double negative population. Gate on the double positive cells, and plot NKp46 versus FSC. Gate on the NKp46 negative population, which are the NKT cells (this gate can be set by finding the point of division in the two populations from the NK cell plot). The NK cells are the tetramer– TCRβ–NKp46+ population. Within NK and NKT cell gates, the IFN-γ+ cells in the PE channel are identified after setting a gate based on the FMO control. Please click here to view a larger version of this figure.
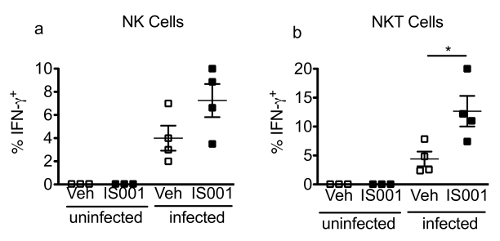
Figure 4. Representative Data Obtained for the Frequencies of IFN-γ+ NK and NKT Cells at 24 hr Post-infection. In this experiment, male C57BL/6J mice (N = 3 – 4/group) were infected i.p. with 105 CFU of L. monocytogenes or were left un-infected. Mice were also administered the drug IS001 or vehicle (0.5% carboxymethyl cellulose) at the same time of inoculation and 12 hr later. Twenty-four hours after inoculation, mice were euthanized and the spleens were removed and were processed individually and stained for flow cytometry. Shown are the mean ± SEM frequency of IFN-γ+ cells in the NK (a) or NKT cell gates (b) in uninfected or infected mice after treatment with a vehicle or the drug IS001. *Indicates a difference (P < 0.05) from vehicle control by two-tailed T-test. Data are re-printed from31 with permission from the Journal of Immunology (volume 195, pp. 5189-5202, 2015). Copyright 2015. The American Association of Immunologists, Inc. Please click here to view a larger version of this figure.
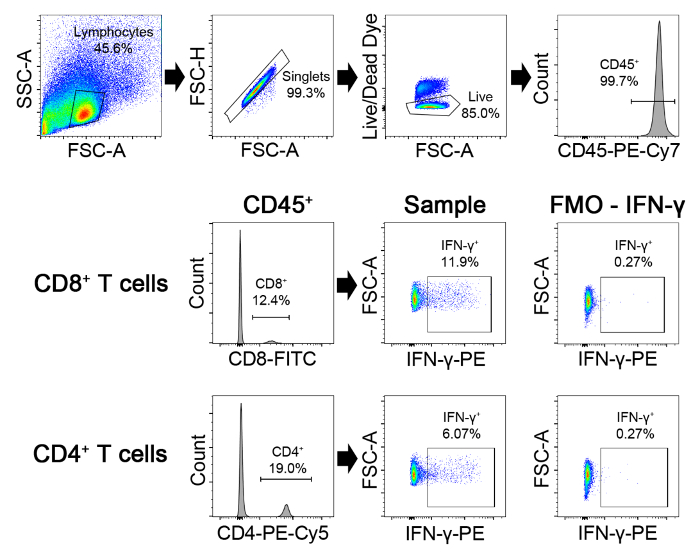
Figure 5. Gating Strategy for Detection of IFN-γ Production in CD4 and CD8 cells. First gate on lymphocytes on FSC-A by SSC-A plots. Then gate on those events that are on the diagonal on the FSC-H/FSC-A plot. These are the singlets. Within this gate, gate on live (AmCyan–) CD45+ cells. Then gate on either CD8+ or CD4+ populations. Within each gate, the IFN-γ+ cells in the PE channel are identified by comparing the staining to the FMO control. Please click here to view a larger version of this figure.
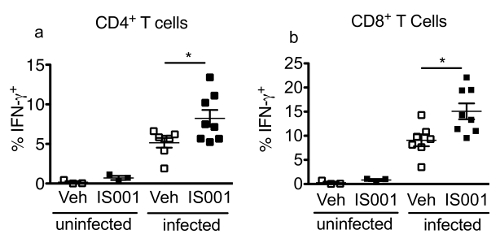
Figure 6. Representative Data Obtained for the Frequencies of IFN-γ+ CD4+ and CD8+ T Cells at 7 Days Post-infection with L. monocytogenes (EGD strain). In this experiment, male C57BL/6J mice were infected i.p. with 2 x 104 CFU L. monocytogenes (N= 7/group) or were left uninfected (N= 3/group). Mice were also administered the drug IS001 or vehicle (0.5% carboxymethyl cellulose) twice daily starting on the day of inoculation. Seven days later, mice were euthanized and the spleens were removed and were processed individually for cell culture. Splenocyte mononuclear cells were stimulated for 24 hr with heat-killed L. monocytogenes with protein transport inhibitor added for the final 4 hr of culture. Cells were then stained for flow cytometry. Shown are the mean ± SEM frequency of IFN-γ+ cells in the CD4+ (a) or CD8+ cell gates (b) in uninfected or infected mice after treatment with a vehicle (0.5% carboxymethyl cellulose) or the drug IS001. * indicates a difference (P < 0.05) from the vehicle control counterpart as determined by two-tailed T test. Data are re-printed from31 with permission from the Journal of Immunology (volume 195, pp. 5189-5202, 2015).Copyright 2015. The American Association of Immunologists, Inc. Please click here to view a larger version of this figure.
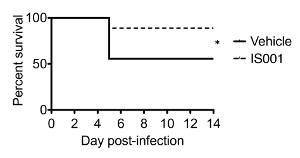
Figure 7. Representative Data Obtained during an Experiment that Compared the Effect of a PPARα Antagonist 1S001 on Mouse Survival to Endpoints after Infection of Male C57BL/6J Mice with L. monocytogenes (EGD strain). In this experiment, male C57BL/6J mice (N = 10 mice/group) were infected i.p. with the modified LD50 dose of the pathogen (105 CFU) of L. monocytogenes. Mice were also administered the drug IS001 or vehicle (0.5% carboxymethyl cellulose) twice daily starting on the day of inoculation. Mice were followed daily for clinical signs and were euthanized if humane endpoints were met. Shown is the percent survival of mice to endpoints over time * indicates a difference in the survival between groups as determined by log-rank test (P < 0.05). Data are re-printed from31 with permission from the Journal of Immunology (volume 195, pp. 5189-5202, 2015). Copyright 2015. The American Association of Immunologists, Inc. Please click here to view a larger version of this figure.
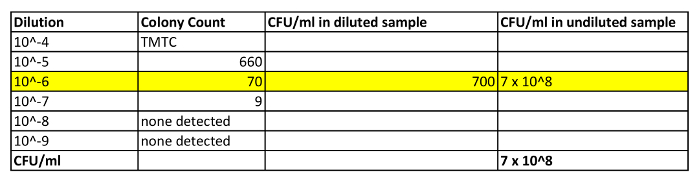
Table 1: Shows Some Representative Calculations for Determining CFU in an Aliquot of Day Culture. In this example, an aliquot of day culture was taken and was diluted 1:1 with BHI media. The OD600 of this diluted sample was determined to be 0.84. In addition, a 100 µl aliquot was taken for CFU determination. This sample was diluted with 900 µl of BHI media (10-1) and was washed and resuspended in 1 ml BHI. A 10-fold dilution series of this sample was prepared (10-2 to 10-9) and diluted samples were plated on BHI agar plates (only values for 10-4 to 10-9 are shown). The next day colonies were counted. Only those plates that had colony numbers between 30-300 were considered for the calculation (i.e., 10-6 plate, highlighted in yellow). The number of colonies on this plate (70) was then divided by 0.1 (volume in ml plated) to get the CFU/ml of the diluted sample. This value was then multiplied by the dilution factor (106) to obtain the CFU/ml reading of the undiluted culture. TMTC = too many to count.
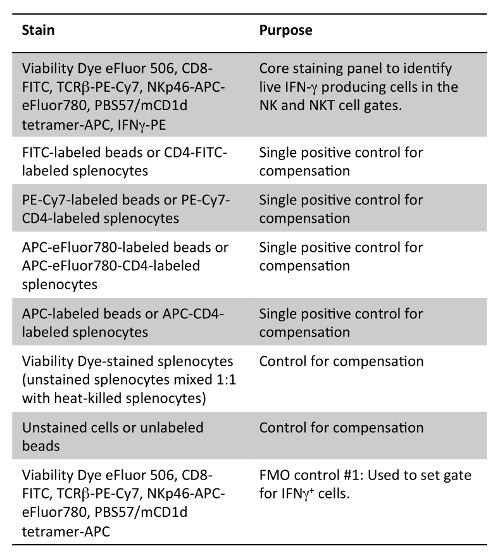
Table 2: Staining Panel for Detection of IFN-γ in NK and NKT Cells. Note that either compensation beads stained with the flow antibodies used in the panel or splenocytes stained with various fluorochrome versions of CD4 antibody clone GK1.1 can be used as single positive controls.
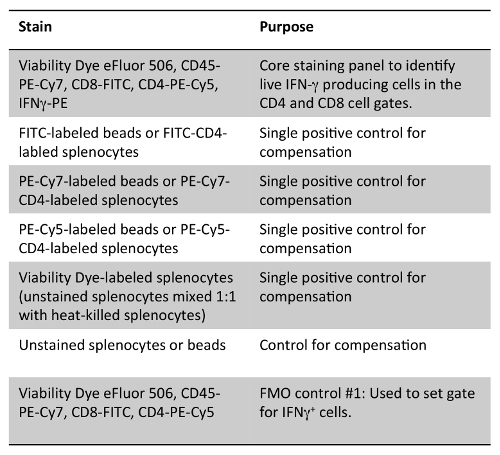
Table 3: Staining Panel for Detection of IFN-γ in CD4+ and CD8+ Cells. Note that either compensation beads stained with the flow antibodies used in the panel or splenocytes stained with various fluorochrome versions of CD4 antibody clone GK1.1 can be used as single positive controls.
Discussion
Here we describe a protocol of how to carry out a basic experimental infection with the EGD strain of L. monocytogenes25 in male or female C57BL/6J mice. This protocol was set up for the purpose of studying the effect of a novel small molecule IS001 on IFN-γ production by innate and adaptive lymphocytes in vivo31. By monitoring bacterial clearance and survival post-infection, insights were gained into how these changes in IFN-γ impacted the host's ability to control the infection.
Critical Considerations in the Protocol
An important consideration in the design of this type of study is that each experiment be adequately powered and appropriately controlled. Due to biological variation in the immune response to infection (see Figures 4 and 6), it is recommended that N = 4 – 5 mice per group should be used for the initial immune studies. If after these studies there is a trend in the data, but no significant difference apparent between groups, a power calculation could be done to determine the least number of animals required in subsequent studies to achieve statistical significance. Regarding controls, it is important to include uninfected controls for determination of baseline IFN-γ responses for immune studies and vehicle controls to help distinguish the effect of the treatment from the stress associated with administering the treatment. Another important consideration is the timing of treatment. Since the innate response to L. monocytogenes is very rapid, it is recommended that the first treatment be administered on the day prior to, or at the same time as, inoculation in order to ensure that therapeutic levels of the reagent be achieved prior to the initiation of the innate immune response.
Yet another important consideration is the dose of the pathogen to be used for infection. A sublethal dose is recommended for measurement of bacterial load, since it increases the chance that the pathogen will be concentrated within the spleen and liver, allowing for the more accurate enumeration of the bacteria. A sublethal dose is also recommended for enumerating IFN-γ responses by adaptive lymphocytes to ensure that animals do not succumb to listeriosis prior to the time of peak T cell expansion. In contrast, it is recommended that a higher infectious dose be used for measurement of the early NK and NKT cell response at 24 hr in order to maximize the IFN-γ production by these cells.
The classical LD50 is the dose of pathogen that results in 50% lethality of mice. Since death was not an acceptable endpoint at our institution and since many symptoms of listeriosis can predict whether an animal is likely to succumb to an infection, we used a defined list of clinical signs instead of death as an endpoint in our studies. Using this method, it was determined that the modified LD50 was 105 CFU for 8-week-old male and 1.5 x 105 CFU for 8-week-old female C57BL/6J mice31. These LD50 doses were determined by measuring the percent survival of mice to endpoints in step-wise dose-escalation studies (N = 5 studies in total) that each contained N = 8 mice per group (e.g., mice were infected first with 10,000 CFU, then a second batch with 20,000 CFU, etc.). The LD50 calculation was determined from a regression plot of the log (CFU) (x-axis) versus the probit of the percent survival values (y-axis) (website: userwww.sfsu.edu/efc/classes/biol710/probit/ProbitAnalysis.pdf).
Note that the modified LD50 dose determined in our lab may differ from that in another lab even when infecting mice with the same strain of L. monocytogenes. Part of this variability may relate to the subjective nature of monitoring clinical signs of listeriosis compared to the more absolute endpoint of death. Additional variability can result from differences in environmental factors such as mouse diet or the microbiota or differences in the preparation of inoculum between labs. Thus, it is recommended that prior to embarking on any survival studies, a pilot study be performed where female mice (N = 8 mice/group) are infected with 1.5 x 105 CFU of the same strain of L. monocytogenes as used in this study and symptoms monitored to determine if this dose indeed results in 50% survival to endpoints. If survival is lower or higher than 50% at this CFU, step-wise dose escalation or dose de-escalation studies could be performed to quickly narrow in on the LD50 dose.
Another important consideration is the strain or substrain of mice used for infection studies. This protocol describes infection of the commonly-used inbred mouse strain C57BL/6J. This strain is well-suited for measurement of IFN-γ responses since this mouse is considered to be a Th1-prone strain33 and as a result, is relatively resistant to L. monocytogenes infection (compared to Th2-prone mouse strains such as BALB/c)34,35. Adapting this protocol to other mouse strains will require knowledge of the infectious dose of the pathogen for the particular strain. It is also recommended to use mice of the same age, sex and vendor as outlined in this protocol in order to reduce the amount of trouble-shooting involved in setting up the model. For example, C57BL/6 mice ordered from one vendor (e.g., C57BL/6J) can exhibit genetic differences than C67BL/6 mice ordered from another vendor (e.g., C57BL/6NTac)36. In addition, the intestinal microbiota differs between C57BL/6 substrains obtained from different vendors, which can influence the balance of Th1 and Th17 responses in the mouse37.
Potential Modifications to Technique
Mice are most commonly inoculated i.p. or intravenously as opposed to the natural route of infection in humans, which is through the gastrointestinal tract. Oral infections are less common because standard strains of L. monocytogenes inefficiently infect the intestinal epithelium of mice38. This is because there is a single amino acid change in the sequence of mouse E-cadherin from human E-cadherin that results in loss of recognition of E-cadherin by the listerial invasion protein, internalin A (InIA)39. To overcome this barrier, researchers use mice that are transgenic for human E-cadherin protein or use listeria that have been engineered to express a mutated sequence of InIA (InIAmut) that binds to mouse E-cadherin with the same affinity as WT EGD for human E-cadherin40. Thus, one potential modification of this technique is to infect mice via the oral route. The reader is referred to another JoVE publication that describes oral inoculation methods38. Note that altering the mode of infection will affect the infectious dose as well as the kinetics of dissemination of the pathogen.
This protocol describes using heat-killed listeria to elicit IFN-γ production by CD4+ and CD8+ T cells. Heat-killed L. monocytogenes was chosen as a stimulus in our studies, because this antigen is inexpensive and because our lab was primarily interested in CD4+ T cell responses to the pathogen. One limitation is that heat-killed bacteria do not efficiently prime CD8+ T cell responses either in vitro41 or in vivo42,43 infection. Thus, the CD8+ T cell IFN-γ production that we observed by splenocytes harvested at the peak of infection (i.e., Figure 6) likely is in response to the residual live bacteria present in the splenocyte cultures or was elicited as a result of cytokine-induced cytokine release41. As an alternative to heat-killed listeria, one could also elicit IFN-γ responses ex vivo by exposing T cells to peptides encoding epitopes on listerial proteins. Indeed, immunodominant MHC Class II-restricted epitopes for listeriolysin O and the p60 hydrolase and have been described for C57BL/6 and BALB/C mice and immunodominant MHC Class I epitopes have been described for BALB/c44. Yet another approach is to infect mice with strains of L. monocytogenes that have been engineered to express model antigens such as ovalbumin or viral antigens in order to take advantage of existing MHC Class I- and MHC Class II-tetramer reagents to enumerate antigen-specific T cells in infected mice45,46.
Other Limitations of the Protocol
Another limitation of this model is that it only measures IFN-γ production by immune cells in the spleen. In addition to the use of tetramers to enumerate antigen specific T cells (of ovalbumin-expressing variants of L. monocytogenes), flow cytometry staining panels described here could be easily modified to measure the production of other cytokines such as TNF or IL-2 or effector molecules that participate in CD8 T cell or NK-mediated killing of the pathogen such as perforin or granzyme B. In addition, this protocol could also be adapted to examine IFN-γ produced by immune cells in the liver.
Future Applications
Once this protocol is mastered, it can serve as a simple in vivo model to screen the effects of various agents or genes on Th1 and cellular immunity.
Divulgations
The authors have nothing to disclose.
Acknowledgements
Development of this protocol was supported by an operating grant from CIHR (MOP97807) to SED.
Materials
| Brain Heart Infusion Broth, Modified | BD | 299070 | any brand should be appropriate |
| Agar | BD | 214010 | any brand should be appropriate |
| Triton X-100 | Sigma-Aldrich | X100 | any brand should be appropriate |
| 1xPBS | Sigma | D8537 | any brand should be appropriate |
| TissueLyser II | Qiagen | 85300 | any brand should be appropriate |
| Ammonium Chloride (NH3Cl) | any brand should be appropriate | ||
| KHCO3 | any brand should be appropriate | ||
| Na2EDTA | any brand should be appropriate | ||
| RPMI 1640 | Gibco | 22400089 | any brand should be appropriate |
| Fetal Bovine Serum | Gibco | 12483 | Before use, heat-inactivate at 56 °C for 30 min |
| L-glutamine | Gibco | 25030 | any brand should be appropriate |
| Non-essential amino acids | Gibco | 11140 | any brand should be appropriate |
| Penicillin/Streptomycin | Gibco | 15140 | any brand should be appropriate |
| GolgiStop Protein Transport Inhibitor (containing Monensin) | BD | 554724 | Use 4 μl in 6 ml cell culture |
| 16% Paraformaledehye | Electron Microscopy Sciences | 15710 | Dilute to 4% PFA in ddH20 or 1xPBS |
| 10 x Perm/Wash buffer | BD | 554723 | Dilute 10x in ddH20 |
| Fc block, Anti-Mouse CD16/CD32 Purified | eBioscience | 14-0161 | Dilute 1:50 |
| Fixable Viability Dye eFluor 506 | eBioscience | 65-0866 | Dilute 1:1000 (we have also used viability dyes from Molecular Probes) |
| anti-Mouse CD4-PE-Cy5 (GK1.5) | eBioscience | 15-0041 | Manufacturer recommends a certain test size; however this should be titrated before use. |
| anti-Mouse CD8-FITC (53-6.7) | eBioscience | 11-0081 | Manufacturer recommends a certain test size; however this should be titrated before use. |
| PBS57/mCD1d tetramer-APC | NIH Tetramer Core Facility | N/A | Obtained as a gift from the facility |
| anti-Mouse TCRβ-PE-Cy7 (H56-597) | eBioscience | 25-5961 | Manufacturer recommends a certain test size; however this should be titrated before use. |
| anti-Mouse NKp46-APC-eFluor780 (29A1.4) | eBioscience | 47-3351 | Manufacturer recommends a certain test size; however this should be titrated before use. |
| anti-Mouse CD45 PE-Cyanine7 (30-F11) | eBioscience | 25-0451 | Manufacturer recommends a certain test size; however this should be titrated before use. |
| anti-Mouse IFN gamma-PE (XMG1.2) | eBioscience | 12-7311 | Manufacturer recommends a certain test size; however this should be titrated before use. |
| OneComp eBeads | eBioscience | 01-1111 | Manufacturer recommends a certain test size; however this should be titrated before use. |
| Mouse IFN gamma ELISA kit | eBioscience | 88-7314 | Used for measuring the interferon gamma in the culture supernatant |
| 50 mL vented tubes for culture | Used for culturing the bacteria, any brand should be appropriate | ||
| 1.5 ml microcentrifuge tubes | any brand should be appropriate | ||
| bacterial petri dishes | any brand should be appropriate | ||
| 2 ml cyrovials | any brand should be appropriate | ||
| UV spectrometer | any brand should be appropriate | ||
| safety engineered needles | any brand should be appropriate | ||
| C57BL6/J | Jackson laboratories | Stock#000664 | Order for arrival at 7 wks |
| Bleach | For decontamination | ||
| 70% Ethanol | For decontamination | ||
| Glass beads | any brand should be appropriate | ||
| Centrifuge | rotor, buckets, bucket covers. | ||
| Microcentrifuge | any brand should be appropriate | ||
| Sterile Glycerol | any brand should be appropriate | ||
| Pipette Tips | any brand should be appropriate | ||
| Pipette | any brand should be appropriate | ||
| Surgical instruments | any brand should be appropriate | ||
| 70 micron strainers | any brand should be appropriate | ||
| 3 ml syringe | any brand should be appropriate | ||
| Pipette gun | any brand should be appropriate | ||
| Filtration Units | any brand should be appropriate | ||
| Trypan Blue | Dilute 1 to 9 in ddH20, any brand should be appropriate | ||
| Hemocytometer | any brand should be appropriate | ||
| Round bottomed plates | any brand should be appropriate | ||
| FACs tubes | BD | ||
| BD LSR II | BD | Any flow cytometer could be used for acquisition that has an appropriate laser configuration and filter set to discriminate the fluorochormes | |
| Flowjo software | Treestar | Used for data analysis. Other types of data analysis software will also be appropriate | |
| Multichannel pipettor (0-300 µl) | Eppendorf | Used for washing cells and adding antibodies during flow cytometry staining | |
| Acetic Acid | Used for washing glass beads, any brand should be appropriate | ||
| Microbank Bacterial Preservation System | Pro-lab Diagnositics | Used as an alternative to glycerol stocks for long-term storage of bacteria |
References
- Pamer, E. G. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 4 (10), 812-823 (2004).
- Ranson, T., et al. Invariant V alpha 14+ NKT cells participate in the early response to enteric Listeria monocytogenes infection. J Immunol. 175 (2), 1137-1144 (2005).
- Bancroft, G. J., Schreiber, R. D., Unanue, E. R. Natural immunity: a T-cell-independent pathway of macrophage activation, defined in the scid mouse. Immunol Rev. 124, 5-24 (1991).
- Soudja, S. M., et al. Memory-T-cell-derived interferon-gamma instructs potent innate cell activation for protective immunity. Immunity. 40 (6), 974-988 (2014).
- Schoenborn, J. R., Wilson, C. B. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. 96, 41-101 (2007).
- Filipe-Santos, O., et al. Inborn errors of IL-12/23- and IFN-gamma-mediated immunity: molecular, cellular, and clinical features. Semin Immunol. 18 (6), 347-361 (2006).
- Flynn, J. L., et al. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 178 (6), 2249-2254 (1993).
- Cooper, A. M., et al. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 178 (6), 2243-2247 (1993).
- Dalton, D. K., et al. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 259 (5102), 1739-1742 (1993).
- Harty, J. T., Bevan, M. J. Specific immunity to Listeria monocytogenes in the absence of IFN gamma. Immunity. 3 (1), 109-117 (1995).
- Huang, S., et al. Immune response in mice that lack the interferon-gamma receptor. Science. 259 (5102), 1742-1745 (1993).
- Wang, Z. E., Reiner, S. L., Zheng, S., Dalton, D. K., Locksley, R. M. CD4+ effector cells default to the Th2 pathway in interferon gamma-deficient mice infected with Leishmania major. J Exp Med. 179 (4), 1367-1371 (1994).
- Hess, J., Ladel, C., Miko, D., Kaufmann, S. H. Salmonella typhimurium aroA- infection in gene-targeted immunodeficient mice: major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J Immunol. 156 (9), 3321-3326 (1996).
- Ikeda, H., Old, L. J., Schreiber, R. D. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 13 (2), 95-109 (2002).
- Hodge, D. L., et al. IFN-gamma AU-rich element removal promotes chronic IFN-gamma expression and autoimmunity in mice. J Autoimmun. 53, 33-45 (2014).
- Seery, J. P., Carroll, J. M., Cattell, V., Watt, F. M. Antinuclear autoantibodies and lupus nephritis in transgenic mice expressing interferon gamma in the epidermis. J Exp Med. 186 (9), 1451-1459 (1997).
- Peng, S. L., Moslehi, J., Craft, J. Roles of interferon-gamma and interleukin-4 in murine lupus. J Clin Invest. 99 (8), 1936-1946 (1997).
- Savinov, A. Y., Wong, F. S., Chervonsky, A. V. IFN-gamma affects homing of diabetogenic T cells. J Immunol. 167 (11), 6637-6643 (2001).
- Miller, C. H., Maher, S. G., Young, H. A. Clinical Use of Interferon-gamma. Ann N Y Acad Sci. 1182, 69-79 (2009).
- Paget, C., Chow, M. T., Duret, H., Mattarollo, S. R., Smyth, M. J. Role of gammadelta T cells in alpha-galactosylceramide-mediated immunity. J Immunol. 188 (8), 3928-3939 (2012).
- Leonard, J. P., et al. Effects of single-dose interleukin-12 exposure on interleukin-12-associated toxicity and interferon-gamma production. Blood. 90 (7), 2541-2548 (1997).
- Zenewicz, L. A., Shen, H. Innate and adaptive immune responses to Listeria monocytogenes: a short overview. Microbes Infect. 9 (10), 1208-1215 (2007).
- Viegas, N., et al. IFN-gamma production by CD27(+) NK cells exacerbates Listeria monocytogenes infection in mice by inhibiting granulocyte mobilization. Eur J Immunol. 43 (10), 2626-2637 (2013).
- Selvanantham, T., et al. Nod1 and Nod2 enhance TLR-mediated invariant NKT cell activation during bacterial infection. J Immunol. 191 (11), 5646-5654 (2013).
- Becavin, C., et al. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. MBio. 5 (2), e00969-e00914 (2014).
- Jones, G. S., D’Orazio, S. E. F. Unit 9B.2 Listeria monocytogenes: cultivation and laboratory maintenance, Chapter 31B. Curr Protoc Microbiol. , (2013).
- Conour, L. A., Murray, K. A., Brown, M. J. Preparation of animals for research–issues to consider for rodents and rabbits. ILAR J. 47 (4), 283-293 (2006).
- Obernier, J. A., Baldwin, R. L. Establishing an appropriate period of acclimatization following transportation of laboratory animals. ILAR J. 47 (4), 364-369 (2006).
- Zhang, M. A., Ahn, J. J., Zhao, F. L., Selvanantham, T., Mallevaey, T., Stock, N., Correa, L., Clark, R., Spaner, D., Dunn, S. E. Antagonizing peroxisome proliferator-activated receptor alpha (PPARalpha) activity enhances Th1 immunity in male mice. J. Immunol. , (2015).
- Kaplan, E. L., Meier, P. Nonparametric estimation from incomplete observations. J Amer Stat Assoc. 53, 457-481 (1958).
- Brown, D. R., et al. Limited role of CD28-mediated signals in T helper subset differentiation. J Exp Med. 184 (3), 803-810 (1996).
- Czuprynski, C. J., Brown, J. F. The relative difference in anti-Listeria resistance of C57BL/6 and A/J mice is not eliminated by active immunization or by transfer of Listeria-immune T cells. Immunology. 58 (3), 437-443 (1986).
- Busch, D. H., Vijh, S., Pamer, E. G. Animal model for infection with Listeria monocytogenes, Chapter 19. Curr Protoc Immunol. , (2001).
- Mekada, K., et al. Genetic differences among C57BL/6 substrains. Exp Anim. 58 (2), 141-149 (2009).
- Ivanov, I. I., et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 4 (4), 337-349 (2008).
- Bou Ghanem, N. E., Myers-Morales, T., Jones, G. S., D’Orazio, S. E. Oral transmission of Listeria monocytogenes in mice via ingestion of contaminated food. J Vis Exp. (75), e50381 (2013).
- Lecuit, M., Dramsi, S., Gottardi, C., Fedor-Chaiken, M., Gumbiner, B., Cossart, P. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. Embo J. 18 (14), 3956-3963 (1999).
- Wollert, T., et al. Extending the host range of Listeria monocytogenes by rational protein design. Cell. 129 (5), 891-902 (2007).
- Brunt, L. M., Portnoy, D. A., Unanue, E. R. Presentation of Listeria monocytogenes to CD8+ T cells requires secretion of hemolysin and intracellular bacterial growth. J Immunol. 145 (11), 3540-3546 (1990).
- Muraille, E., et al. Distinct in vivo dendritic cell activation by live versus killed Listeria monocytogenes. Eur J Immunol. 35 (5), 1463-1471 (2005).
- Datta, S. K., et al. Vaccination with irradiated Listeria induces protective T cell immunity. Immunity. 25 (1), 143-152 (2006).
- Geginat, G., Schenk, S., Skoberne, M., Goebel, W., Hof, H. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J Immunol. 166 (3), 1877-1884 (2001).
- Shen, H., et al. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc Natl Acad Sci U S A. 92 (9), 3987-3991 (1995).
- Foulds, K. E., et al. Cutting edge: CD4 and CD8 T cells are intrinsically different in their proliferative responses. J Immunol. 168 (4), 1528-1532 (2002).

