Bidirectional Retroviral Integration Site PCR Methodology and Quantitative Data Analysis Workflow
Summary
This manuscript describes the experimental procedure and software analysis for a bidirectional integration site assay that can simultaneously analyze upstream and downstream vector-host junction DNA. Bidirectional PCR products can be used for any downstream sequencing platform. The resulting data are useful for a high-throughput, quantitative comparison of integrated DNA targets.
Abstract
Integration Site (IS) assays are a critical component of the study of retroviral integration sites and their biological significance. In recent retroviral gene therapy studies, IS assays, in combination with next-generation sequencing, have been used as a cell-tracking tool to characterize clonal stem cell populations sharing the same IS. For the accurate comparison of repopulating stem cell clones within and across different samples, the detection sensitivity, data reproducibility, and high-throughput capacity of the assay are among the most important assay qualities. This work provides a detailed protocol and data analysis workflow for bidirectional IS analysis. The bidirectional assay can simultaneously sequence both upstream and downstream vector-host junctions. Compared to conventional unidirectional IS sequencing approaches, the bidirectional approach significantly improves IS detection rates and the characterization of integration events at both ends of the target DNA. The data analysis pipeline described here accurately identifies and enumerates identical IS sequences through multiple steps of comparison that map IS sequences onto the reference genome and determine sequencing errors. Using an optimized assay procedure, we have recently published the detailed repopulation patterns of thousands of Hematopoietic Stem Cell (HSC) clones following transplant in rhesus macaques, demonstrating for the first time the precise time point of HSC repopulation and the functional heterogeneity of HSCs in the primate system. The following protocol describes the step-by-step experimental procedure and data analysis workflow that accurately identifies and quantifies identical IS sequences.
Introduction
Retroviruses insert their genomic DNA into the host genome at various sites. This unique property, which may contribute to the development of cancers and other forms of viral pathogenesis, has the ironic benefit of making these viruses highly amenable to cellular engineering for gene therapy and basic biology research. The viral Integration Site (IS) – the location on the host genome where a foreign DNA (virus) is integrated – has important implications for the fate of both the integrated viruses and the host cells. IS assays have been used in various biological and clinical research settings to study retroviral integration site selection and pathogenesis, cancer development, stem cell biology, and developmental biology1,2,3,4. Low detection sensitivity, poor data reproducibility, and frequent cross-contamination are among the key factors limiting the applications of IS assays to current and planned studies.
Many IS analysis technologies have been developed. Restriction enzyme-based integration site assays, including Linker-Mediated (LM) Polymerase Chain Reaction (PCR)5, inverse PCR6, and Linear-Amplification-Mediated (LAM) PCR7, are the most widely used. The use of site-specific restriction enzymes, however, generates a bias during the retrieval of the IS, allowing only a subset of integromes (a foreign DNA integrated into the host genome) in the vicinity of the restriction site to be recovered4. Assay technologies that more comprehensively assess vector IS have also been introduced in recent years. These assays employ various strategies, including Mu transposon-mediated PCR8, nonrestrictive (nr)-LAM PCR9, type-II restriction enzyme-mediated digestion10, mechanical shearing11, and random hexamer-based PCR (Re-free PCR)12, to fragment genomic DNAs and amplify IS. Current technologies have varying levels of detection sensitivity, genome coverage, target specificity, high-throughput capacity, complexity of assay procedures, and biases in detecting the relative frequencies of target sites. Given the varying qualities of the existing assays and the variety of purposes for which they can be used, the optimal assay approach should be carefully selected.
This work provides detailed experimental procedures and a computational data analysis workflow for a bidirectional assay that significantly improves detection rates and sequence quantification accuracy by simultaneously analyzing the IS upstream and downstream of the integrated target DNA (see Figure 1 for a schematic view of the assay procedures). This approach also provides the means to characterize the retroviral integration process (for example, the fidelity of target site duplication and variations in the genomic sequences of upstream and downstream insertions). Other bidirectional methods have been used primarily for cloning and sequencing both ends of the target DNA11,13,14. This assay is extensively optimized for the high-throughput and reproducible quantification of vector-marked clones, using the well-established LM-PCR method and computational analysis mapping, and for quantifying both upstream and downstream junctions. Bidirectional analysis with the TaqαI enzyme has proven useful for high-throughput clonal quantification in stem cell gene therapy preclinical studies2,15. This paper describes a modified method using a more frequent cutter (RsaI/CviQI – motif: GTAC) that doubles the chances of detecting integromes compared to a TaqαI-based assay. Detailed experimental and data analysis procedures that use GTAC motif enzymes for lentiviral (NL4.3 and its derivatives) and gamma-retroviral (pMX vectors) vector IS analysis are described. The oligonucleotides used in the assay are listed in Table 1. An in-house programming script for IS sequence analysis is provided in the supplemental document.
Protocol
1. Generating Upstream (left)- and Downstream (right)-junction Sequence Libraries
- DNA linker preparation:
- Prepare a 10 µL linker DNA solution by adding 2 µL of 100 µM LINKER_A oligos (final: 20 µM), 2 µL of 100 µM LINKER_B oligos (final: 20 µM), 2 µL of 5 M NaCl (final: 1M), and 4 µL of nuclease-free water in a PCR tube. See Table 1 for the linker sequences.
- Incubate the linker DNA solution at 95 °C for 5 min in a PCR instrument, stop the run program, and turn the PCR instrument power off. Leave the linker solution in the instrument for 30 min to slowly cool down the linker DNA. The linker DNA solution can be stored at 4 °C.
- LTR-specific biotin-primer extension:
- Use a UV-Vis spectrophotometer to determine the DNA concentration and 260/280 nm values of the genomic DNA from in vivo or in vitro experiments. Dilute 1-2 µg of sample genomic DNA to a final volume of 170 µL of genomic DNA solution using nuclease-free water.
- Prepare a 200 µL PCR reaction by mixing 2.5 µL each of 10 µM HIV-1-specific biotin primers (L-BPs and R-BPs in Table 1: total of 10 µL for four lentivirus-specific primers), 20 µL of 10X thermostable DNA polymerase buffer, 4 µL of 10 mM dNTPs (final: 200 µM of each dNTP), 3 µL of 2.5 U/µL thermostable DNA polymerase, and 163 µL of genomic DNA.
NOTE: For gammaretroviral vectors (pMX vectors), use 5 µL each of two 10 µM pMX-specific biotin primers (L-BP and R-BP: total of 10 µL) instead of the four HIV-1-specific biotin primers above. - Divide the solution into four PCR tubes (each 50 µL) and carry out a single extension cycle under the following condition: 94 °C for 5 min, 56 °C for 3 min, 72 °C for 5 min, and 4 °C for storage.
- Pool all four PCR reactions into a 2 mL microcentrifuge tube and follow the PCR purification procedure of the PCR purification kit. Elute with 50 µL of elution buffer (5-fold water-diluted elution buffer provided in the kit). Immediately proceed with step 1.3.1 or store the eluted DNA at -20 °C.
- RsaI and CviQI digestion
- Prepare a 100 µL digestion reaction by adding 50 µL of DNA (from step 1.2.4), 10 µL of 10x buffer A, and 2 µL (20 U) of RsaI enzyme. Incubate at 37 °C for 1 h in a PCR instrument.
- Add 1 µL (10 U) of CviQI enzyme to the reaction and incubate at 25 °C for 30 min in a PCR instrument. Immediately proceed with step 1.4.1
- Blunt ending:
- Prepare a 4.5-µL mixture containing 2.5 µL of DNA Polymerase I large (klenow) fragment and 2 µL of 10 mM dNTPs. Transfer 1 µL of the mixture to the DNA sample from step 1.3.2. The total volume will be 102 µL. Mix well by vortexing and incubate at 25 °C for 1 h in a PCR instrument. Immediately proceed with step 1.6.1.
- Preparation of streptavidin beads:
- Briefly vortex the streptavidin bead solution and transfer 50 µL to a new 2 mL microcentrifuge tube. Remove the supernatant using the magnetic stand and wash the beads with 200 µL of binding solution.
- Resuspend the beads in 100 µL of binding solution and place the tube away from the magnetic stand. Immediately proceed with step 1.6.1.
- Streptavidin bead binding:
- Transfer 100 µL of sample DNA (step 1.4.1) to the 100 µL re-suspended bead solution (step 1.5.2) and mix carefully by pipetting to avoid any foaming of the solution. Incubate the tube at room temperature for 3 h on a rotating wheel or a roller.
- Use the magnetic stand to capture the beads and discard the supernatant. Wash the beads twice in 400 µL of washing solution and twice in 400 µL of 1x T4 DNA ligase buffer (diluted from 10X solution using nuclease-free water).
- Resuspend the beads in 200 µL of 1x T4 DNA ligase buffer (diluted from 10X solution using nuclease-free water) and place it away from the magnetic stand. Immediately proceed with step 1.7.1.
- Linker ligation:
- Prepare a 400 µL ligation reaction solution in a 2 mL microcentrifuge tube by mixing 0.5 µL of the DNA linker (step 1.1.2), 10 µL of 10x T4 DNA ligase buffer, 20 µL of 5X T4 DNA ligase buffer (containing 25% polyethylene glycol), 5 µL of T4 DNA ligase, 164.5 µL of nuclease-free water, and 200 µL of the re-suspended beads (step 1.6.3). Place the reaction tube on the rotating wheel and incubate at RT (22 °C) for 3 h (or 16 °C O/N).
- Wash the beads twice with the washing solution and twice with 1X thermostable DNA polymerase buffer (diluted from 10x solution using nuclease-free water) using the magnetic stand.
- Resuspend the beads in 50 µL of 1x thermostable DNA polymerase PCR buffer and place it away from the magnetic stand. Immediately proceed with step 1.8.1 or store at 4 °C for up to one day.
- Pre-amplification of both the left and right junction DNA:
- Prepare a 200 µL PCR reaction by adding 10 µL of 10 µM 1L-primer, 10 µL of 10 µM 1R-primer, 20 µL of 10 µM primer Link1 (Table 1), 10 µL of 10X thermostable DNA polymerase buffer, 4 µL of 10 mM dNTPs (final: 200 µM of each dNTP), 8 µL (20 U) thermostable DNA polymerase, and 88 µL of nuclease-free water to the re-suspended beads (50 µL) from step 1.7.3.
- Aliquot the reaction mixture into 4 PCR tubes (each 50 µL) and carry out PCR with the following condition: 94 °C incubation for 2 min; 25 cycles of 94 °C for 20 s, 56 °C for 25 s, and 72 °C for 2 min; and a final extension at 72 °C for 5 min.
- Pool all 4 PCR reactions into a 2 mL microcentrifuge tube and follow the PCR purification procedure of the PCR purification kit. Elute with 50 µL of elution buffer.
- Determine the DNA concentration and 260/280-nm values using a UV-Vis spectrophotometer; the DNA can be stored at -20 °C until ready for the next step. Proceed with steps 1.9.1/1.10.1 (optional) or directly with steps 1.11.1/1.12.1.
- Removing the left-side internal DNA amplicon (optional):
- Transfer up to 100 ng of PCR DNA product from step 1.8.4 to a 2 mL microcentrifuge tube and adjust the volume to 10 µL using nuclease-free water.
- Prepare a restriction enzyme reaction specifically targeting the left-side internal DNA amplicon.
NOTE: The reaction condition may differ depending on the choice of enzyme. For example, when removing the left-side internal DNA from NL4.3-based lentiviral vectors, add 1 µL of pvuII, 2 µL of buffer B, and 7 µL of nuclease-free water. Incubate at 37 °C for 1 h in a PCR instrument. Immediately proceed with step 1.11.1 or store at -20 °C.
- Removing the right-side internal DNA amplicon (optional):
- Proceed the same as in step 1.9.1.
- Prepare a restriction enzyme reaction specifically targeting the right-side internal DNA amplicon.
NOTE: For example, when removing the right-side internal DNA from NL4.3-based lentiviral vectors, add 1 µL of sfoI, 2 µL of buffer B, and 7 µL of nuclease-free water. Incubate at 37 °C for 1 h in a PCR instrument. Immediately proceed with step 1.12.1 or store at -20 °C.
- Left-junction-specific amplification:
- Prepare a 50 µL PCR reaction by mixing 5 µL of DNA from step 1.8.4 (or from the optional step 1.9.2), 5 µL of 10 µM 2L-primer (final: 1 µM), 5 µL of 10 µM primer Link2 (final: 1 µM), 5 µL of 10x thermostable DNA polymerase buffer, 1 µL of 10 mM dNTPs (final: 200 µM of each dNTP), 2 µL (5 U) of thermostable DNA polymerase, and 27 µL of nuclease-free water.
- Carry out the PCR with the following condition: 94 °C incubation for 3 min; 8-15 cycles of 94 °C for 20 s, 56 °C for 25 s, and 72 °C for 2 min; and a final extension at 72 °C for 5 min.
NOTE: Amplified DNA may be stored at -20 °C. *Cycle number optimization is suggested. - Follow the PCR purification procedure of the PCR purification kit. Elute with 50 µL of elution buffer. Determine the DNA concentration and 260/280 nm values.
- Right-junction-specific amplification:
- All procedures are identical to "Left-junction-specific amplification" (steps 1.11.1-1.11.3), except for the use of different primers; use the right-junction-specific primer (2R-primer, see Table 1) in this step.
- PCR amplicon length variation test:
- Analyze the PCR amplicon length variations by performing 2% agarose gel electrophoresis or capillary electrophoresis (Figure 2).
NOTE: This is an essential step to confirm the completion of the assay procedures and to make a rough assessment of IS patterns based on the PCR band patterns. The purified PCR amplicon from steps 1.11.3 and 1.12.3 can be used for various DNA sequencing platforms. Proceed with the appropriate sample preparation procedures for classical chain-termination (Sanger) sequencing or next-generation sequencing. The DNA may be stored at -20 °C.
- Analyze the PCR amplicon length variations by performing 2% agarose gel electrophoresis or capillary electrophoresis (Figure 2).
2. Computational IS Sequence Analysis
- Preparation of data files:
- Prepare three input data files: a fasta format sequence data file (Test_data.fa in supplemental data), a tsv file for the search motifs for vector and linker sequences (Demultiplexing_Trimming_blunt_GTAC.tsv in supplemental data), and a tsv file for restriction enzyme information (Enzyme.tsv in supplemental data).
NOTE: These files are required for demultiplexing, trimming vector and linker sequences, and removing internal vector sequences (Figure 3A). Detailed step-by-step instructions for implementing the computational workflow are provided in the README.txt file in the supplemental data.
- Prepare three input data files: a fasta format sequence data file (Test_data.fa in supplemental data), a tsv file for the search motifs for vector and linker sequences (Demultiplexing_Trimming_blunt_GTAC.tsv in supplemental data), and a tsv file for restriction enzyme information (Enzyme.tsv in supplemental data).
- Computational analysis:
- Run demultiplexing and trimming scripts.
NOTE: The raw sequence will be processed for demultiplexing and for the trimming of the vector, linker, and primer sequences (see STEP-1 in the supplemental README.txt file). - Run the mapping command (see STEP-2 in the README.txt file) to map the processed sequences onto the reference genome using a BLAST-like alignment tool (BLAT; www.genome.ucsc.edu).
- Run the quantitative IS analysis script (see STEP-3 in the README.txt file).
NOTE: Two output files (Initial_count_without_homopolymer_correction.txt and Final_count.txt) will be generated. More details on mapping and sequence enumeration strategies can be found in the "README.txt file in the supplemental data and in previous publications2,15.
- Run demultiplexing and trimming scripts.
Representative Results
The bidirectional IS assay generated different sizes of PCR amplicons for both the upstream (left) and downstream (right) vector host junctions (Figure 2). The size of a PCR amplicon is dependent on the location of the nearest GTAC motif upstream and downstream from an integrome. The assay also produced internal DNA PCR amplicons: retroviral sequences near the polypurine tract and the primer binding site were concomitantly amplified during left- and right-junction PCR, respectively. PCR amplicon bands can be visualized by capillary or agarose (2%) electrophoresis.
After sequencing, both the left- and right-junction sequences were analyzed by an in-house programming script for preprocessing raw sequences (including demultiplexing and trimming vector and linker DNA), mapping IS sequences onto the genome, identifying and counting identical IS sequences, and applying error correction procedures (Figure 3). Locations of the 5'-end of the query sequences in the reference genome are considered IS and are used for initial counting of each IS. The criteria determining the two matching sequences-the upstream (left) and downstream (right) junction sequences originated from the same integrome-are based on the nucleotide sequence patterns of retrovirus-specific concerted integration and are as follows: (i) Two junction sequences align onto the genome in th opposite orientations. (ii) The IS of the two junction sequences are separated by a 5-bp overlap for human immunodeficiency virus (HIV) vectors and a 4-bp overlap for murine leukemia virus (MLV) vectors. The first 5 bp of the two junctions of HIV and 4 bp of MLV junctions are reverse-complementary.
IS sequence counts are determined in three steps: (1) mapping IS sequences onto the genome using BLAT, which generates the mapping quality and separates individual IS sequences into "single-hit," "multi-hit," "no-hit," and "others" groups; (2) using the Basic Local Alignment Search Tool (BLAST) to compare single-hit sequences with other suboptimally mapped sequences, including multi-hits, no-hits, and others; and (3) identifying and correcting sequencing errors, including homopolymer errors. Multi-hit sequences are IS sequences that can align two or more genomic positions with a high mapping score. While still useful for identifying and quantifying clonal populations, the multi-hit sequences cannot be used for characterizing genomic integration site distribution patterns (for example, association with genes, repeats, or other genomic characters). In some rare cases, the two junction sequences show different mapping qualities. For example, one shows "single-hit," while the matching sequence shows "multi-hit." In such cases, both sequences are treated as "single-hit" sequences.
A portion of IS sequences with suboptimal mapping scores, showing high percent genome matching (identity) with an abnormal query size (QSIZE), or vice versa, were separated from "no-hits" and grouped into "others" for an additional and often manual re-evaluation. For example, when using BLAT for genome mapping, some IS sequences may show an abnormal QSIZE due to miss-matching nucleotides in the first or last 5-10 nucleotides. These sequences often do not meet the mapping criteria for "single-hit" or "multi-hit" status, despite having a relatively high-quality mapping result.
A sample raw sequence data file (Test_DATA.fa: 33,374 sequences) and sample output data files are provided as supplementary data. 1 µg of genomic DNA from human repopulating cells, transduced with lentiviral vectors (FG12) in a humanized bone marrow/liver/thymus (BLT) mouse17, were analyzed using the bidirectional assay. Retroviral IS were found all over the genome. Typically, lentiviral vectors are overrepresented in genes, whereas gamma-retroviral vectors are overrepresented in transcription start sites16 (Figure 4). From two human repopulating cell samples, a total of 1,081 sequences-851 from the upstream (left) and 230 the downstream (right) junctions-were qualified as IS sequences. From these sequences, 93 unique IS in the left and 50 unique IS in the right junctions were identified. Of these, 44 were identified in both (left and right) junctions, showing a total of 99 unique integromes in the test samples. IS are significantly enriched in genes (66%, p <0.0001) compared to random events (Figure 4A).
Sample gamma-retrovirus vector (pMX) integration site sequences are also included in the test data file. From a mixed Pmx-engineered cell sample, transduced with pMX expressing Oct4, cMyc, Klf4, and Sox2, 1,611 IS sequences and 129 unique IS were identified. Of 65 and 76 unique IS identified in the left and right junctions, respectively, 12 were in both junctions.
It has been previously shown that PCR amplicons of ≥500 bp are poorly sequenced in the pyrosequencing platform, whereas PCR amplicons of <500 bp are generally well-sequenced, without a notable bias with regards to sequence lengths15. Thus, sequence data from ≥ 500 bp PCR amplicons were excluded to remove length-associated sequencing bias. Only data from <500 bp PCR amplicon (termed as quantifiable vector integrome, or QVI) were used for quantitative clonal analysis. The relative detection frequencies of vector integromes were calculated using only the sequence counts of IS junctions generating <500 bp PCR amplicons (Figure 4B-4D; also see Table 2). Approximately 77% of the vectors could be quantitatively analyzed by this strategy (Figure 4B). As a result, the calculated frequencies for each QVI were expected to over-estimate the true frequencies in the sample (Figure 4E) by 1.25x.
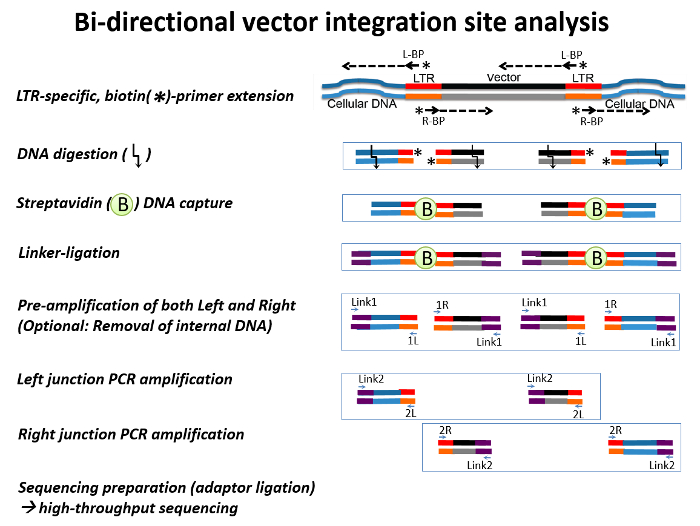
Figure 1: A Schematic View of Bidirectional Integration Site Analysis. Double-stranded retroviral DNA (black and red) flanked by cellular DNA (blue) are shown. The arrows represent oligonucleotide primers, and arrows with an asterisk represent biotin primers. Linker DNAs are denoted by purple lines. Briefly, a linear extension of left biotin primers (L-BP) and right biotin primers (R-BP) from the viral Long Terminal Repeat (LTR) generates biotinylated, double-strand IS DNA. After digestion with CviQI and RsaI, the biotinylated double-stranded DNA are enriched using streptavidin-biotin-specific binding and are ligated with linker DNA. Streptavidin-captured, linker-ligated vector-host junction DNAs are amplified by a two-step PCR: pre-amplification (amplification of both the left and right junctions) followed by two nested PCRs, each targeting left- and right-junctions. Please click here to view a larger version of this figure.
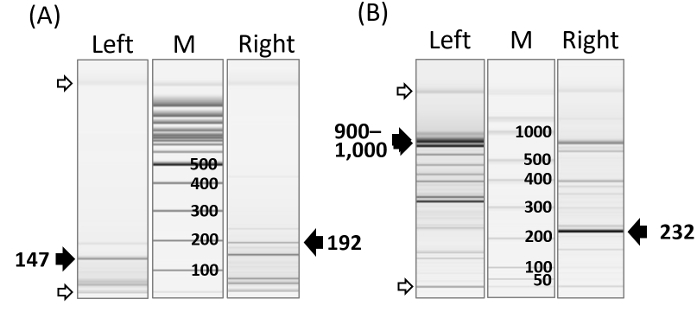
Figure 2: Representative PCR Amplicon Image. (A) Capillary electrophoresis analysis shows varying lengths of PCR amplicons in lentiviral vector (FG12) integration sites after pvuII or sfoI digestion. Varying PCR bands for upstream (left) and downstream (right) vector host junctions are shown. The dark arrow heads indicate the internal vector DNA amplicons remaining after pvuII or sfoI digestion. The DNA size marker (0.1 - 2.5 kbp) is on the M lane. The open arrow heads indicate alignment markers (15 & 5,000 bp). DNA alignment markers are used for the calibration of the migration time variation across all channels. (B) Gamma-retroviral (pMX) vector integration sites in murine cell clones transduced with multiple pMX vectors. Left- and right-junction DNAs, as well as internal vector DNA amplicons (arrow heads), are shown. The dark and open arrow heads indicate internal vector DNA and alignment markers, respectively. The DNA size marker (50-1,500 bp) is on the M lane. Details on capillary electrophoresis can be found in the company protocol. Please click here to view a larger version of this figure.
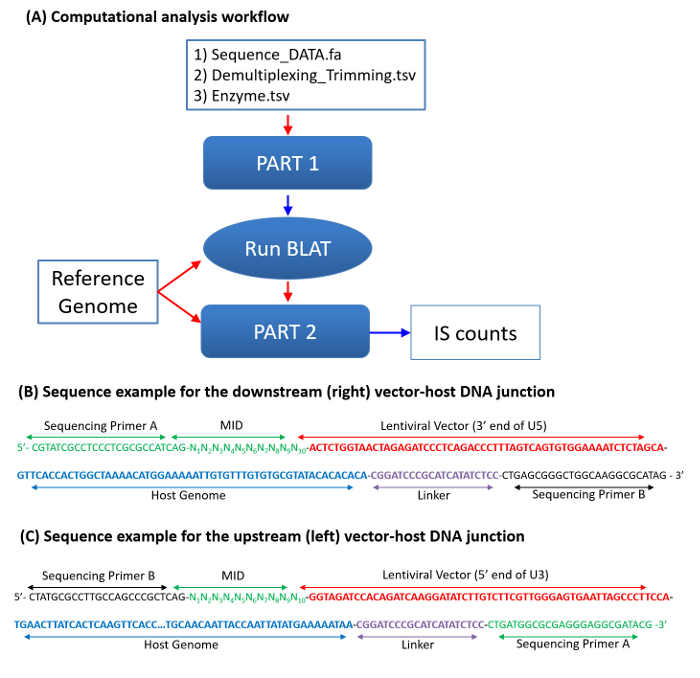
Figure 3: Integration Site Sequence Analysis. (A) A flowchart for computational data analysis. Three data files, including a fasta format sequence file, a file with reference sequence motifs for demultiplexing and trimming, and a file with restriction enzyme information, are required. Sample files, including Test_DATA.fa (sequence data), Demultiplexing_Trimming.tsv (search sequence motifs), and Enzyme.tsv (restriction enzyme recognition sequence), are provided as supplemental data files. The processed sequences (host-genome sequences) from Part 1 will be mapped against the reference using BLAT. Locations at the 5'-end of the query sequences in the reference genome are considered to be the integration sites (Table 2). Sequence counts for each IS are determined in three steps: (1) mapping IS sequences onto the genome using BLAT; (2) comparing single-hit IS sequences (aligning onto a unique site of the host genome) with other sequences that were suboptimally mapped onto the genome; and (3) identifying and correcting sequencing errors. More details on mapping and sequence enumeration strategies can be found in previous studies2,15. (B-C) Two sample single-stranded DNA sequences for the downstream (B) and the upstream (C) vector-host junctions are shown. Pyrosequencing primers A and B (green) are used for droplet PCR and sequencing. The color codes agree with those in Figure 1. The sample downstream junction sequence (B) includes Primer A (green), MID (green), Vector U5 end (red), host genome (blue), linker (purple), and Primer B (green). In a mixed (upstream and downstream) DNA sequencing, Primer A is used for sequencing the downstream junctions (B), and Primer B is used for sequencing the upstream junctions. The sample upstream junction sequence (C) includes Primer B, MID, Vector U3 end, host genome, linker, and Primer A. Please click here to view a larger version of this figure.
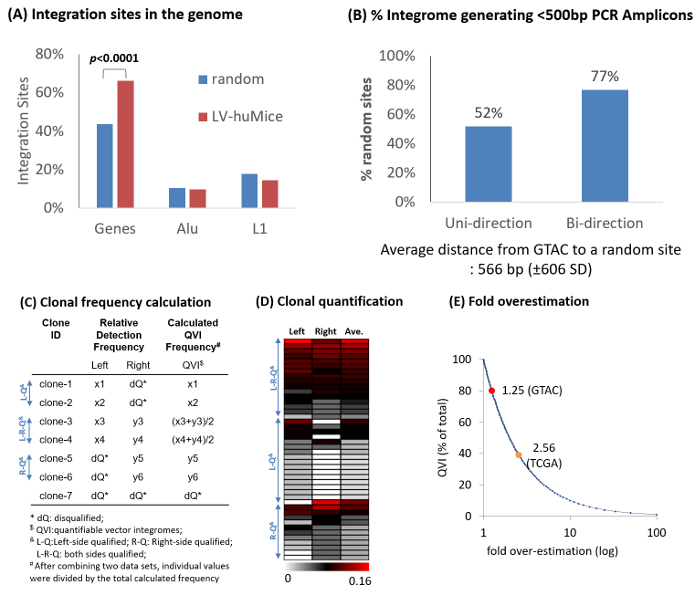
Figure 4: Representative Example of Bidirectional Integration Site Analysis. (A) Percent integration in Genes (refseq), Alu, and LINE 1 (L1) repeats of lentiviral vectors in humanized mouse repopulating cells (LV-huMice), in comparison with in silico-generated 10,000 random integration events. Integration sites are mapped onto the human genome (hg19). LV-huMice integration sites are significantly over represented in Genes (p <0.0001, chi-square approximation) (B) In silico analysis of 10,000 random integration events. With a unidirectional approach, approximately 52% of random integromes generated PCR amplicons of <500 bp, whereas with a bidirectional approach, 77% generated a PCR amplicon of <500 bp in either the left or right junctions. PCR amplicons longer than 500 bp are inefficiently sequenced with the pyrosequencing platfor 2,15 and should thus be excluded from quantitative data analyses. (C) Strategy for clonal quantification. Each individual clone shares the same vector integrome (or IS). The relative frequencies of the left (x) and right (y) junctions are combined to represent the relative quantities of the clonal populations (quantifiable vector integromes, or QVI). Integration sites that do not have a GTAC motif within 450 bp are disqualified (dQ) and removed from quantification analysis. (D) The relative frequencies (relative to all QVI sequences) of 44 QVI clones in humanized mice repopulating cells are shown in a color scheme (white to red: 0 to 0.16). (E) Expected over-estimation of clonal frequencies with bidirectional analysis. Based on in silico 10,000 random integration analysis, a 1.25-fold over-estimation is expected when using GTAC motif enzymes (RsaI and CviQI) because of approximately 20% dQ clones. A 2.56x over-estimation is expected because of approximately 60% dQ clones when using the TCGA motif enzyme (TaqαI). Please click here to view a larger version of this figure.

Table 1: Oligonucleotides for Lentiviral and Gamma-retroviral Vector Integration Site Analysis. * 5BioTEG: Biotin modification at the 5' end. Please click here to view a larger version of this table.
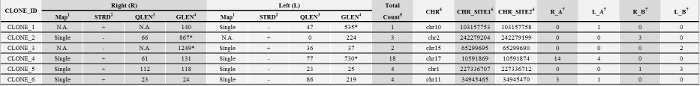
Table 2: Representative Insertion Site Sequence Count Data. Please click here to view a larger version of this table.
1 Map: integration site sequences can be mapped onto a unique location (single-hit) or multiple loci (multi-hit) of the reference genome. N.A. (not available): no sequence was detected.
2 STRD: orientation of the query sequence in the genome
3 QLEN: the length of the query sequence
4 GLEN: the expected length of the integration site sequence, calculated based on the distance from the nearest available GTAC motif in the genome to the insertion site. GLEN <450 bp are accepted for quantitative analyses.
5 Total_Count: total sequence count
6 Integration sites are shown by a chromosome number (CHR) and a site number for the right junction (CHR_SITE1) and a number for the left junction (CHR_SITE2). CHR_SITE1 and CHR_SITE2 are 5 bp apart for lentiviral vectors and 4 bp apart for MLV (pMX) vectors
7 Sequence counts for the right junction (R_A) and the left junction of sample A(L_A); sequence counts for the right junction (R_B) and the left junction of sample B(L_B)
* Disqualified (dQ): GLEN ≥450 in silico bp
Discussion
The bidirectional assay enables the simultaneous analysis of both the upstream (left) and downstream (right) vector-host DNA junction sequences and is useful in a number of gene therapy, stem cell, and cancer research applications. The use of GTAC-motif enzymes (RsaI and CviQI) and the bidirectional PCR approach significantly improves the chances of detecting an integrome (or a clonal population) when compared to previous TCGA-motif enzyme (TaqαI)-based assays2,15 and other unidirectional LM-PCR approaches5. The bidirectional assay improves the analysis of IS, particularly in the limited DNA samples that often arise in clinical or small-animal-model studies.
Steps 1.1-1.7 are critical step and should be done without unnecessary delays. These steps are usually done in one day. Testing enzymes prior to these steps is highly recommended. Steps 1.9 and 1.10 are optional. These steps will reduce internal vector DNA sequences that are concomitantly amplified with IS sequences. Table 1 provides primer sets suitable for analyzing the IS of wildtype NL4.3 HIV-1, NL4.3-derived vectors, including FG1221, and pMX-based gammaretroviral vectors22. Depending on the nucleotide variations in the long terminal repeat (LTR) sequences, the primer design and experimental approach may need a proper modification.
The blat software used for mapping is only compatible with fasta format and does not work with fastq files. One can convert the fastq files to fasta using various software tools, such as FASTX-Toolkit or BBTools. A user with basic knowledge of python can use Biopython to convert the fastq files to fasta for mapping them with blat.
It is expected that a portion of the IS will not be detected with the bidirectional IS assay and, even if detected, will not qualify for downstream clonal quantification. When GTAC-motif enzymes were used, approximately 23% of the integromes in the sequence data generated by the pyrosequencing platform did not pass the analysis criteria for quantitative IS sequence analysis (Figure 4). When comprehensive IS coverage is critical, for example in the safety monitoring of gene-engineered cells in gene therapy settings, it is advisable to choose an assay with unrestricted genome access to IS 8,9,10,11,12 or to to reanalyze the same sample with a different or optimized combination of restrictases4,18.
IS assays with unrestricted genome access, such as non-restrictive LAM PCR and random-shearing approaches19,20, hold particular promise for comprehensive IS analysis. These approaches use technologies that are relatively difficult to control, making it hard to predict the outcome of genomic DNA fragmentation and IS amplification. On the other hand, IS assays using well-characterized restriction enzymes have two major benefits: (1) It is relatively easy to calibrate and optimize assays, because the assay outcomes are more predictable due to the specificity of the enzyme reaction and the availability of reference genome sequences. (2) Sequence data are highly reproducible once the assay conditions have been optimized. The bidirectional PCR with optimized conditions has proven useful for large-scale and accurate clonal quantification2,15. Although only a portion of existing clonal populations have been analyzed due to restriction enzyme bias, the quantities of individual clones were accurately measured, thereby enabling the accurate determination of clone size variations within and across samples. A sufficient number of clones were generated to determine stem cell behavior patterns and functional heterogeneity.
The PCR amplicons produced from this bidirectional PCR procedure are suitable for any downstream sequencing platform. Due to the high sensitivity of the assay, the utmost care should be taken to prevent cross-contamination by performing experiments in a contamination-free room. The inclusion of a negative (no-template) control experiment is advised for all PCR steps. Even with the most careful practice, preventing sample-to-sample cross-contamination is extremely difficult. Thus, when comparing clonal populations from different samples, it is advisable to employ a collision control, which removes potential contaminated data23, and a cutoff for low-frequency, unreliable clones2 in order to minimize noise from cross-contaminated DNA. The bidirectional approach generates IS data for both ends of the integromes, thereby providing an additional opportunity to reduce potential false-positive detection errors.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Funding was provided by the National Institutes of Health Grants R00-HL116234, U19 AI117941, and R56 HL126544; the National Science Foundation Grant DMS-1516675; the National Research Foundation of Korea (NRF-2011-0030049, NRF-2014M3C9A3064552); and the KRIBB initiative program.
Materials
| Thermostable DNA polymerase | Agilent | 600424 | PicoMaxx Polymerase |
| Thermostable DNA polymerase buffer | Agilent | 600424 | PicoMaxx Polymerase buffer |
| Deoxynucleotide (dNTP) solution mix | New England Biolabs | N0447L | dNTP solution mix (10mM each) |
| PCR tubes | VWR International | 53509-304 | PCR Strip Tubes With Individual Attached Caps |
| 2ml microcentrifuge tube | Molecular Bioproducts | 3453 | microcentrifuge tubes |
| PCR purification kit | Qiagen | 28106 | |
| RsaI | New England Biolabs | R0167L | restriction enzyme |
| CviQI | New England Biolabs | R0639L | restriction enzyme |
| Buffer A | New England Biolabs | B7204S | NEB CutSmart buffer |
| DNA Polymerase I large (klenow) fragment | New England Biolabs | M0210L | Blunting |
| streptavidin beads solution | Invitrogen | 60101 | Dynabeads kilobaseBINDER kit |
| Binding Solution | Invitrogen | 60101 | Dynabeads kilobaseBINDER kit |
| Washing Solution | Invitrogen | 60101 | Dynabeads kilobaseBINDER kit |
| magnetic stand | ThermoFisher | 12321D | DynaMag™-2 Magnet |
| T4 DNA ligase | New England Biolabs | M0202L | T4 DNA ligase |
| 10X T4 DNA ligase buffer | New England Biolabs | B0202S | T4 DNA ligase reaction buffer |
| 5X T4 DNA ligase buffer | Invitrogen | 46300-018 | T4 DNA ligase buffer with polyethylene glycol-8000 |
| UV-Vis spectrophotometer | Fisher Scientific | S06497 | Nanodrop 2000 |
| pvuII | new England Biolabs | R0151L | restriction enzyme |
| sfoI | new England Biolabs | R0606L | restriction enzyme |
| Buffer B | new England Biolabs | B7203S | NEB buffer 3.1 |
| Nuclease free water | Integrated DNA Technologies | 11-05-01-14 | |
| Capillary electrophoresis | Qiagen | 9001941 | QIAxcel capillary electrophoresis |
| Veriti 96-well Fast Thermal Cycler | Thermo Fisher Scientific | 4375305 | PCR Instrument |
| Rotating wheel (or Roller) | Eppendorf | M10534004 | Cell Culture Roller Drums |
| DNA size marker | Qiagen | 929559 | QX size marker (100-2,500 bp) |
| DNA size marker | Qiagen | 929554 | QX size marker (50-1,500 bp) |
| DNA alignment markers | Qiagen | 929524 | QX DNA Alignment Marker |
| genomc DNA | Not Available | Not Available | Sample genomc DNA from in vivo or in vitro experiments |
References
- Serrao, E., Engelman, A. N. Sites of Retroviral DNA Integration: From Basic Research to Clinical Applications. Crit. Rev. Biochem. Mol. Bio. 51 (1), 26-42 (2016).
- Kim, S., et al. Dynamics of HSPC Repopulation in Nonhuman Primates Revealed by a Decade-Long Clonal-Tracking Study. Cell Stem Cell. 14 (4), 473-485 (2014).
- Bushman, F. Retroviral integration and human gene therapy. J. Clin. Invest. 117 (8), 2083-2086 (2007).
- Bystrykh, L. V., Verovskaya, E., Zwart, E., Broekhuis, M., de Haan, G. Counting stem cells: methodological constraints. Nat Meth. 9 (6), 567-574 (2012).
- Schröder, A., et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 110 (4), 521-529 (2002).
- Silver, J., Keerikatte, V. Novel use of polymerase chain reaction to amplify cellular DNA adjacent to an integrated provirus. J. Virol. 64, 3150 (1990).
- Schmidt, M., et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR). Nat. Meth. 4 (12), 1051-1057 (2007).
- Brady, T., et al. A method to sequence and quantify DNA integration for monitoring outcome in gene therapy. Nucleic Acids Res. 39 (11), e72 (2011).
- Gabriel, R., et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat. Med. 15 (12), 1431-1436 (2009).
- Kim, S., Kim, Y., Liang, T., Sinsheimer, J., Chow, S. A high-throughput method for cloning and sequencing human immunodeficiency virus type 1 integration sites. J. Virol. 80 (22), 11313-11321 (2006).
- Maldarelli, F., et al. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 345 (6193), 179-183 (2014).
- Wu, C., et al. High Efficiency Restriction Enzyme-Free Linear Amplification-Mediated Polymerase Chain Reaction Approach for Tracking Lentiviral Integration Sites Does Not Abrogate Retrieval Bias. Hum. Gene. Ther. 24 (1), 38-47 (2013).
- Aker, M., Tubb, J., Miller, D. G., Stamatoyannopoulos, G., Emery, D. W. Integration Bias of Gammaretrovirus Vectors following Transduction and Growth of Primary Mouse Hematopoietic Progenitor Cells with and without Selection. Mol. Ther. 14 (2), 226-235 (2006).
- Gabriel, R., Kutschera, I., Bartholomae, C. C., von Kalle, C., Schmidt, M. Linear Amplification Mediated PCR; Localization of Genetic Elements and Characterization of Unknown Flanking DNA. J Vis Exp. (88), e51543 (2014).
- Kim, S., et al. High-throughput, sensitive quantification of repopulating hematopoietic stem cell clones. J. Virol. 84 (22), 11771-11780 (2010).
- Mitchell, R., et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2 (8), e234 (2004).
- Melkus, M., et al. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat. Med. 12 (11), 1316-1322 (2006).
- Bystrykh, L. V. A combinatorial approach to the restriction of a mouse genome. BMC Res Notes. 6, 284 (2013).
- Beard, B. C., Adair, J. E., Trobridge, G. D., Kiem, H. -. P. High-throughput genomic mapping of vector integration sites in gene therapy studies. Methods Mol Biol. , 321-344 (2014).
- Gabriel, R., et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat. Med. 15 (12), 1431-1436 (2009).
- Qin, X., An, D., Chen, I., Baltimore, D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A. 100 (1), 183-188 (2003).
- Kitamura, T., et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31 (11), 1007-1014 (2003).
- Cartier, N., et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 326 (5954), 818-823 (2009).

