Visualization and Live Imaging of Oligodendrocyte Organelles in Organotypic Brain Slices Using Adeno-associated Virus and Confocal Microscopy
Summary
Myelinating oligodendrocytes promote rapid action potential propagation and neuronal survival. Described here is a protocol for oligodendrocyte-specific expression of fluorescent proteins in organotypic brain slices with subsequent time-lapse imaging. Further, a simple procedure for visualizing unstained myelin is presented.
Abstract
Neurons rely on the electric insulation and trophic support of myelinating oligodendrocytes. Despite the importance of oligodendrocytes, the advanced tools currently used to study neurons, have only partly been taken on by oligodendrocyte researchers. Cell type-specific staining by viral transduction is a useful approach to study live organelle dynamics. This paper describes a protocol for visualizing oligodendrocyte mitochondria in organotypic brain slices by transduction with adeno-associated virus (AAV) carrying genes for mitochondrial targeted fluorescent proteins under the transcriptional control of the myelin basic protein promoter. It includes the protocol for making organotypic coronal mouse brain slices. A procedure for time-lapse imaging of mitochondria then follows. These methods can be transferred to other organelles and may be particularly useful for studying organelles in the myelin sheath. Finally, we describe a readily available technique for visualization of unstained myelin in living slices by Confocal Reflectance microscopy (CoRe). CoRe requires no extra equipment and can be useful to identify the myelin sheath during live imaging.
Introduction
The brain's white matter is composed of nerve cell axons wrapped in myelin, a specialized extended plasma membrane formed by oligodendrocytes. Myelin is required for fast and reliable action potential propagation and long-term survival of myelinated axons, and a loss of myelin can cause neurological dysfunction. Despite their importance, the properties of oligodendrocytes are less known compared with neurons and astrocytes. Consequently, fewer tools have been developed for studying oligodendrocytes.
Live imaging of cell organelles such as mitochondria, endoplasmatic reticulum (ER) or different vesicular structures can be useful to study dynamic changes in the organelles over time. Traditionally, imaging of living oligodendrocytes has been performed in monocultures1,2. However, oligodendrocytes in monoculture do not display compact myelin, and organotypic or acute brain slices may, therefore, be a better option when studying localization and movement of organelles. Localization of small organelles and proteins in the myelin sheath can be challenging due to the short distance between the myelinated axon and the surrounding myelin sheath. Thus, light microscopic immunostaining procedures alone do not have the spatial resolution to discriminate between organelles in the myelin sheath and those in the myelinated axon. This can be solved by viral transduction with genes for organelle-targeted fluorescent proteins driven by cell type-specific promoters. The advantages are a cell-specific and sparse expression, which enables accurate assessment of organelle localization and dynamics. Transgenic animals can also be used to achieve such an organelle-targeted cell-specific expression3. However, the production and maintenance of transgenic animals is expensive and usually does not offer the sparse expression that can be achieved by viral methods.
The method described here uses viral transduction of oligodendrocytes with mitochondrial-targeted fluorescent proteins (dsred or green fluorescent protein, GFP) driven by the myelin basic protein promoter (MBP-mito-dsred or MBP-mito-GFP) to visualize oligodendrocyte mitochondria in organotypic brain slices. In addition, expression of another fluorescent protein in the cytoplasm (either GFP used together with mito-dsred or tdtomato used with mito-GFP) is used to enable visualization of cell morphology, including the cytoplasmic compartments of the myelin sheath. The protocol includes the procedure for making organotypic brain slices (a modified version of the protocol described by De Simoni and Yu, 20064,5). We then describe the time-lapse imaging procedure for studying mitochondrial movement. This procedure uses an upright confocal microscope with a continuous exchange of imaging medium, a setup that enables easy application of drugs or other medium changes during imaging. The time-lapse imaging procedure can be performed on any confocal microscope, with some extra equipment for maintaining living slices as described below. The protocol also contains several tips to optimize imaging and reduce phototoxicity.
Lastly, a quick and simple way to visualize unstained myelin by Confocal Reflectance microscopy (CoRe) is described. This can be useful to identify the myelin sheath during live imaging. In recent years, several techniques have been developed to image myelin without any staining required, but most of these require specific equipment and expertise6,7,8. The procedure described here uses the reflective properties of the myelin sheath and is a simplified single-excitation wavelength version of Spectral Confocal Reflectance microscopy (SCoRe, in which several laser wavelengths are combined to visualize myelin)9. CoRe can be done on any confocal microscope that has a 488 nm laser and a 470 – 500 nm bandpass emission filter or a tunable emission filter.
Protocol
The procedures described here have been approved by the Norwegian Animal Research Authority. Suppliers and catalog numbers for the consumables and other required equipment are available in the materials list at the end of the document.
1. Preparation of Organotypic Slices
NOTE: This recipe uses two mouse pups at postnatal day 7-9 (p7-9), which yield 24 organotypic slices divided on two six-well culture dishes. Unless stated otherwise, all procedures should be done in a sterile hood and nitril or latex gloves should be used. Only cell culture grade ingredients should be used.
- Autoclave bottles, dissection tools (1 large scissor, 1 small scissor, 1 large flat spatula, 1 small rounded and sharpened spatula and 1 forceps, see Materials list for details), glass pipettes, pipette tips and tissue wipes used for the slice preparation.
- As half of the glass pipettes should be used with the large opening, break off the narrow end.
- Score with a diamond scriber pen (if available) around the pipette just below the point where the glass starts narrowing. Then place the pointy end of the pipette into a nitrile glove or similar. Carefully break off the tip by bending the pipette while pushing it against a work bench.
- Then blunt the broken end by rubbing on a piece of sand paper or melt slightly on a Bunsen burner. The broken pipettes are used to transfer cut brain slices with the broken end turned into the rubber bulb and the large open end touching the solution/slices.
NOTE: This is not done in a sterile hood and can be done several days in advance.
- Prepare culture dishes.
NOTE: This can be done the day before slicing or at least 2 hours before slicing.- Sterilize Polytetrafluoroethylene (PTFE) membranes ("confetti"). Use two sterile forceps to remove single confetti from the surrounding blue plastic pieces and sterilize by dipping twice into a Petri dish containing 96% ethanol (EtOH). Then lay confetti flat in a sterile large Petri dish to dry.
NOTE: The confetti are hydrophilic PTFE membranes similar to the membranes in the culture inserts. The use of confetti aids the live imaging because single slices can be easily transferred from the insert to the microscope setup by lifting the confetti with forceps (see 4.8. for details). Alternatively, the brain slices can be cultured without the confetti (in which the slices will attach directly to the membrane of the culture insert). In this case, the membrane insert must be cut with a scalpel to transfer the slice to the microscope bath. - Add 1 mL of culture medium (recipe in Reagents' table) to each well of the six-well culture dishes.
- Place one culture insert in each well using sterile forceps. Avoid air bubbles between the insert and the culture medium.
NOTE: To save money and the environment, it is possible to reuse the culture inserts. This can be done by thorough rinsing in distilled H2O (dH2O) followed by incubation in 70% EtOH for a minimum of 24 h until reuse. When preparing for new culture, dry the inserts in a cell culture plate under ultraviolet (UV) light for 15-30 min. - Place two (sterilized and dry) confetti on each of the culture inserts. Place the confetti towards the edges of the inserts to get minimal overlap.
- Put the plates in the cell culture incubator at 37 °C with 5% CO2.
- Sterilize Polytetrafluoroethylene (PTFE) membranes ("confetti"). Use two sterile forceps to remove single confetti from the surrounding blue plastic pieces and sterilize by dipping twice into a Petri dish containing 96% ethanol (EtOH). Then lay confetti flat in a sterile large Petri dish to dry.
- Bubble cold dissection medium (recipe in Reagents' table) with bioxide gas (95% O2/5%CO2). To keep the solution sterile, connect the tube from the gas cylinder to a sterile syringe filter (0.22 µm pore size).
- Couple the other end of the syringe filter to a sterile tube connected to a sterile glass pipette. Place the glass pipette in the solution, cover with parafilm and turn on the gas. Keep the dissection medium on ice.
- Prepare the dissection/slicing area.
NOTE: Ideally, this procedure should be done in a sterile hood. If this is not possible, the vibratome can be left on a work bench. In this case, it is recommended to use a hair net and face mask.- Using a single edge razor blade, cut out a rectangular piece (approximately 2 cm x 0.5 cm) of agarose gel (recipe in Reagents' table) and glue this onto the vibratome stage such that the long side faces the vibratome blade.
- Clean all surfaces and the vibratome parts with 70% EtOH and wipe with autoclaved tissue wipes.
NOTE: It is important that all the pieces that will be in contact with the brain tissue are completely dry since EtOH can damage the cells. - Attach a sterile razor blade to the vibratome and perform vibration check if the vibratome has this function.
- Place autoclaved dissection tools in the hood together with autoclaved tissue wipes, four small Petri dishes, a round-edge scalpel, 2 glass pipettes with rubber bulbs, two open-end glass pipettes (see pt. 1.1) and super glue (sprayed with EtOH).
- Pour bubbled dissection medium into the vibratome boat and into the four small Petri dishes.
NOTE: This step should only be done immediately before dissection since medium from this point on is not bubbled or kept on ice.
- Dissect and mount brains.
NOTE: To obtain healthy slices, this step must be done quickly and precisely. Some practice is needed.- Euthanize animal #1 by dislocation of the neckor according to local guidelines and then decapitate with the large scissors.
- Using small, sharp scissors, quickly remove the skin by making a sagittal incision from the neck to the nose and pull aside to reveal the skull.
- Cut along the sagittal suture, followed by lateral incisions over the interior part of the frontal bones and the lamboid suture (the border between cerebrum and cerebellum).
- Use forceps to bend open the skull on each side. The cranial nerves are cut off from the brain by inserting a rounded, sharp spatula under the ventral side of the brain and gently moving the spatula laterally.
- Use a single edge razor blade to make a coronal cut rostral to the cerebellum.
NOTE: This cut will determine the angle at which the slices are cut. It should, therefore, be made at a straight angle. - Flip the brain out of the skull and into a small Petri dish containing dissection medium.
- Repeat steps 1.6.1. – 1.6.4. for animal #2 and place this brain in a second Petri dish.
NOTE: The animals are not sterile. Perform the dissection on one side of the hood and then move to the other, clean side once the brains have been removed from the skull. - Change gloves.
- Attach brain to vibratome stage: add a thin layer of super glue to the stage just in front of the mounted agarose gel. Hold brain #1 with a large flat spatula with the freshly cut (caudal) side touching the spatula and the ventral side facing (and being as close as possible to) the agarose gel.
- Then gently place the brain onto the glue by pushing it off the spatula with a set of forceps. Make sure to leave enough space for brain #2.
NOTE: It is important that the brains are well attached to the stage, but without excessive glue, as this can obstruct the cutting.
- Then gently place the brain onto the glue by pushing it off the spatula with a set of forceps. Make sure to leave enough space for brain #2.
- Immediately after mounting of brain #1, use the large spatula to add some drops of dissection medium on top of the brain. This will keep the brain moist, harden the glue and prevent it from sticking to the sides of the brains.
- Repeat steps 1.6.7 and 1.6.8. for brain #2.
- Attach the stage onto the vibratome boat.
- Cut 230 µm thick coronal slices with an amplitude of 1 – 1.5 mm, a frequency of 85 Hz and a velocity of 0.2 mm/s. Collect slices approximately between 1.4 mm rostral and 2.1 mm caudal to Bregma.
- Collect slices after each slice has been cut by using open-end glass pipettes (pt. 1.1). Transfer slices to a Petri dish containing dissection medium. Use a rounded scalpel to cut the slices into two pieces, dividing the two hemispheres.
- When all the slices are cut, place slices into the culture dishes.
- Take the culture dish (one at the time) out of the incubator.
- Using the open-end glass pipettes, carefully transfer one slice to each of the confetti.
NOTE: The slices should lay flat in the middle of the confetti. This requires some skill. However, if some slices are not perfect, it is better to leave them than to spend time trying to move them as it is important to get all the slices into the incubator as fast as possible. - Use a glass pipette (pointy end) to gently remove excess medium without touching the slice.
NOTE: Slices should not be immersed in liquid during the incubation. Thus, excess medium must be aspirated such that the slices are exposed to the air (a thin film of medium will still cover the slices). Slices that remain immersed in liquid will usually be unhealthy or die (4 and our observations). - Move the dish back in the incubator once all the confetti have one slice.
- Repeat steps 1.7.8. – 1.8.4. for dish #2.
- Change the culture medium.
NOTE: This should be done the day after slicing (day 1 in vitro, DIV1) and then every 2-3 days.- Preheat culture medium in a water bath or the incubator.
- In a sterile hood, use vacuum suction or a regular pipette with a sterile tip to aspirate culture medium from each well. This is done by gently tipping the dish (by about 30 degrees) and placing the pipette tip at the edge of the culture insert.
- Remove approximately 800 µL from each well (leaving just enough medium to keep the bottom of the insert covered in medium). Then, using a clean pipette, add 800 µL of the pre-heated medium to each well. Then place the dishes back in the cell culture incubator.
2. Viral Transduction
- Purchase or make AAVs with a plasmid of interest, as described previously10,11.
NOTE: This protocol describes the use of AAV serotype 2/8 (AAV 2/8) carrying mitochondrial targeted dsred or GFP as a reporter gene under the transcriptional control of the myelin basic protein promoter12 (AAV2/8 MBP_mito_dsred or AAV2/8 MBP_mito_GFP) to visualize oligodendrocyte mitochondria. AAV 2/8 carrying GFP or tdtomato as reporter gene driven by the general CAG promoter (AAV 2/8 CAG_GFP or AAV 2/8 CAG_tdtomato) is used to visualize oligodendrocyte cytoplasm. Thus, the combination of AAV2/8 MBP_mito_dsred with AAV 2/8 CAG_GFP gives red mitochondria and green cytoplasm in oligodendrocytes, whereas combining AAV2/8 MBP_mito_GFP and AAV 2/8 CAG_tdtomato gives green mitochondria and red cytoplasm. The constructs are described in more detail elsewhere13. - On day 7 in vitro (DIV7), transduce oligodendrocytes with the AAVs.
CAUTION: Follow safety regulations for working with AAVs. Make sure to have the proper training and the Biosafety Level approval required. All the following points should be carried out in a Class II biosafety cabinet using gloves and lab coat. Here, application of AAV to the cortex area of the slices is described, as this is the area that shows the best recovery.- Dilute the AAV in pre-heated (37 °C) culture medium. Dilutions around 2 × 109 genome copies/mL (GC/mL) are recommended to get a sparse transduction of oligodendrocytes which enables imaging of single cells within the slice. However, a pilot using different titers should be done to ensure a density of transduced oligodendrocytes that is suitable for the specific experiments. If two different AAVs are used, they should be mixed in the same solution and added to the slice simultaneously.
- Add around 1.3 µL diluted AAV to each slice: Make sure the outside of the pipette tip is dry. Pipette 1.3 µL diluted AAV and hold the pipette above the cortex, as close as possible without touching the slice with the pipette tip (about 1 mm away).
- Then slowly push solution out of the pipette. When the solution touches the slice, move the pipette tip over the cortex to spread the solution over the whole cortex area.
NOTE: This procedure should be done as fast as possible to avoid keeping the slices out of the hood for longer than necessary. Do this on one dish at the time and put the first dish back in the incubator before starting on dish #2. To get a satisfactory distribution of AAV without damaging the tissue requires some practice and a steady hand.
- If an upright fluorescence microscope is available in the cell lab, protein expression can be checked during the time in culture by placing the dish under the microscope.
NOTE: The dish should not be kept out of the incubator for more than 2 – 5 minutes.
3. Time-lapse Imaging
NOTE: The imaging can be performed whenever the expression levels of the fluorescent markers are sufficient and the slices look healthy (usually DIV11-14).
- Ensure that the microscope, perfusion and heating are set up correctly so that bubbled imaging solution is heated and can enter and exit the bath.
NOTE: Various solutions exist for bath chambers. A simple version is presented here.- Make in- and outlets of the bath by blunted syringe needles (bent at ~45°) that are attached to the sides of the bath by blu-tack/fun tack.
- Ensure that tubing goes from a bottle of continuously bubbled imaging solution, via the peristaltic pump, through the heating tube of the heater unit, and then to the inlet syringe needle in the bath.
- Connect another tube to the syringe needle outlet. This tube then goes via the peristaltic pump and back to the bottle of imaging solution (or to a waste bottle).
- Bubble imaging solution (recipe in Reagents' table) with bioxide gas.
NOTE: This should be started at least 15 minutes before imaging and continue throughout the experiment. - Turn on peristaltic pump and heater and run solution through the bath to obtain optimal bath temperature (37 ± 0.5 °C). Ensure that the thermometer sensor connected to the temperature unit is submerged in the bath solution.
- Turn on the microscope, fluorescence lamp and appropriate lasers.
- Set up a suitable light path for the sample.
NOTE: This will vary depending on microscope setup and probe. General knowledge of how to use the confocal microscope is required and will not be dealt with here. - Use a water immersion objective with the appropriate magnification and numerical aperture (n.a.). We use a 40x water immersion plan-apochromat objective (n.a. 1.0).
- Open pinhole to achieve an optical section of approximately 2 – 2.5 µm for the time-lapse video. This reduces the occurrence of mitochondria moving out of focus during time-lapse.
- Transfer organotypic slice to the bath:
- Add a drop of imaging solution or culture medium to a small plastic Petri dish.
- In a sterile hood, use sterile forceps to transfer one organotypic slice from the membrane insert to the drop. The forceps should only touch the confetti and not the slice.
- Put the lid on the Petri dish.
- Immediately put the culture dish containing the rest of the slices back in the incubator.
- Bring Petri dish containing slice to the microscope.
- Turn off peristaltic pump while transferring slice to microscope bath. First, use forceps to lift confetti with slice from Petri dish to the top of the bath (the confetti will float). Then place the two tips of the forceps on each side of the confetti so that they touch the confetti without touching the slice, and push the confetti through the solution to the bottom of the bath. Center the confetti in the middle of the bath.
- Use forceps to place the hold-down anchor (harp) on top of the confetti without touching the slice.
NOTE: A harp is a horseshoe-shaped platinum anchor that is used to prevent the slice from floating or drifting in the bath. For acute slices, thin strings run from one side of the harp to the other (resembling a music harp). For organotypic slices, the harp should be stringless and fit the size of the confetti in such a way that it can lay on top of the confetti without touching the slice. - Turn peristaltic pump back on.
- Lower the objective down to the sample.
- Select cell and region to image.
NOTE: Avoid excessive exposure of the cells to laser light as this can cause cell toxicity (see tips in the points below).- Use the eyepieces and the fluorescent lamp to find a healthy-looking oligodendrocyte with the correct expression. Typically, oligodendrocytes that have a strong overexpression of fluorescent protein appear unhealthy. Unhealthy oligodendrocytes will have processes with a blobby appearance, and the mitochondria will appear fragmented. In healthy oligodendrocytes, the soma and processes appear smooth and mitochondria have various lengths (although typically much shorter than in neurons and astrocytes13,14,15).
- Place the cell of interest in the middle of the field of view.
- Use the "live" function in the software (for Leica and Zeiss microscopes) to identify the cell on the screen and choose an area of interest, e.g., a part of the cell that contains several primary processes or myelin sheaths, or a single process or myelin sheath.
NOTE: To be able to image as much of the processes/sheaths as possible, choose areas containing processes that lay relatively flat in the x-y direction. - Zoom in on the field of interest (typically producing an image with pixel size of 0.14 – 0.30 µm).
- Using the "regions" tool, draw a rectangle around the area and choose the option to scan only the selected region. Scanning time, and thus laser exposure, is reduced by only scanning the selected region.
NOTE: Horizontal rectangular regions will be scanned faster than vertical regions due to the shorter number of scanned lines needed. If a vertical rectangle region is scanned, the scanning time can be reduced by rotating the scan field by 90 degrees (using the rotation tool).
- Adjust laser power and gain to get a good view on mitochondria.
NOTE: Use the minimal laser power needed. If possible, turn up gain rather than increasing laser power. In addition to causing cell toxicity, too high laser power will bleach the imaged objects over time, thus requiring a further increase of laser power or gain during scanning. The required laser power and gain will depend on the expression level and the microscope. With an LSM700 confocal microscope, we use a 555 nm laser at a power of 20-40 µW to image dsred or tdtomato and a 488 nm laser is used at 20 – 30 µW to image GFP (laser power measured at the back aperture of the objective). Gain is set high for live imaging, usually in the range of 700 – 800 for GFP and 850-980 for dsred and tdtomato. The high gain may cause some more background noise, which is removed by increasing the digital offset (offset values typically lay between 0 and 15). In our experience, using MBP_mito_GFP gives a better signal-to-noise than MBP_mito_dsred. - Select scan speed.
NOTE: Minimize scanning time by using a fast scanning speed and by drawing a small scanning region (see point 4.10.5 below). It is also possible to save scanning time by performing a bidirectional scan. However, this is not recommended due to poorer image quality. - Set frequency and duration of time-lapse recording, e.g. capture images every 2 seconds for a total of 20 minutes for mitochondrial imaging in oligodendrocytes.
NOTE: The frequency and duration should be set according to the mobility of the objects of interest. A higher frequency will expose the specimen to more laser light, but faster-moving objects also require a shorter total duration of time-lapse videos (e.g. mitochondria in neurons, which move more frequently and at a much higher speed than mitochondria in oligodendrocytes, are typically imaged every second for a total of 2 minutes16). - Start time-lapse recording.
NOTE: The mitochondria may move out of focus and/or drift out of the imaged region during the recording. To minimize vibrations and movement, adjust in- and outlets of the bath and reduce pump speed if needed. Small changes in focus usually still occur. Thus, continuous monitoring of the screen during imaging is required, with small adjustments of the focus during the recording. - Capture a z-stack of the whole cell for future identification of cell and imaged process.
NOTE: For better resolution, the pinhole should be reset to 1 airy unit for the z-stack. - Optional: Visualize unstained myelin.
NOTE: In oligodendrocytes transduced with (cytoplasmic) GFP, the myelin sheaths can usually be identified as straight lines of cytoplasm (the cytoplasmic ridges) connected to a primary process (see Figure 2 A and B). However, if GFP expression is too weak or a cytoplasmic marker is not used, it is possible to visualize the myelin sheath by confocal reflectance microscopy (CoRe) as explained here.- Set up a new light path in the software with laser excitation at 488 nm and capture emitted light around the same wavelength, e.g. 470-500 nm.
- Adjust laser power and gain until myelin is visible (see example in Figure 3). Using an LSM 510 Meta microscope, we typically use a laser power of 7 - 12 µW (measured at the back aperture of the objective).
- Optional: preparation of slices for immunostaining.
- If the imaged cell must be identified after immunostaining, capture a low magnification image (10x) of the cell and indicate its location in the image by drawing an arrow or similar. To aid detection of the cell, it is recommended to also draw a manual map of the whole slice, indicating the location of the cell.
- Fix slice in 4% paraformaldehyde (PFA) on shaker for 1 hour in room temperature.
CAUTION: PFA is harmful. Use protective wear and only use in a ventilated hood. - Dilute fixative 1:10 in PBS and leave at 4°C until starting immunohistochemistry as described elsewhere17.
NOTE: Although slices can be left in diluted fixative for several weeks, fluorescence may fade. Performing immunohistochemistry within 10 days of fixation is recommended.
Representative Results
Organotypic brain slices that were cultured and transduced as described above showed a sparse distribution of cortical oligodendrocytes expressing mito_dsred and GFP. Immunostaining with antibodies against Olig2 and MBP confirmed that the expression was specific to oligodendrocytes (Figure 1).
For live imaging, transduced oligodendrocytes were recognized by their characteristic morphology of several myelin sheaths running in parallel (Figure 1 and Figure 2A). By performing time-lapse imaging on the transduced oligodendrocytes, it is possible to monitor the movement of mitochondria in primary processes and myelin sheaths (Figure 2B–D).
In cells with low GFP expression, the myelin sheath could be identified during live imaging by illuminating the sample at 488 nm and capturing emitted light 470 – 500 nm (Figure 3A). This Confocal Reflectance microscopy (CoRe) technique also worked on PFA fixed and immunostained tissue, although myelin sheaths appeared dimmer (Figure 3B–C). By immunostaining with antibodies against myelin basic protein (MBP), we found that close to 100% of myelin sheaths laying horizontal to the imaging plane (indicated by arrowheads, Figure 3B–C) are visible with CoRe, whereas myelin sheaths that lay perpendicular to the field of view (indicated by asterisks, Figure 3B–C) are less or not at all visible. Although nearly all internodes are visualized, many internodes appear discontinuous, with small parts of the sheath not being visible (Figure 3A insert). These small "gaps" in the myelin can be filled in by imaging with three complementary wavelengths, so-called SCoRe9.
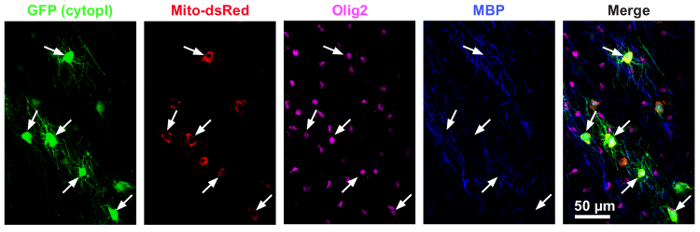
Figure 1: Immunohistochemistry confirms that MBP-mito-dsred is selectively expressed in oligodendrocytes that are immune positive for Olig2 and MBP. Images are from the neocortex of cultured mouse brain slices transduced with (AAV2/8) MBP-mito-dsred (red, mitochondrial marker) and CAG-GFP (green, cytosol). The slices were immunolabeled for the myelin marker Myelin Basic Protein (MBP, blue) and Olig2 (oligodendrocyte lineage cell nuclear marker, magenta). (Figure modified from reference13) Please click here to view a larger version of this figure.
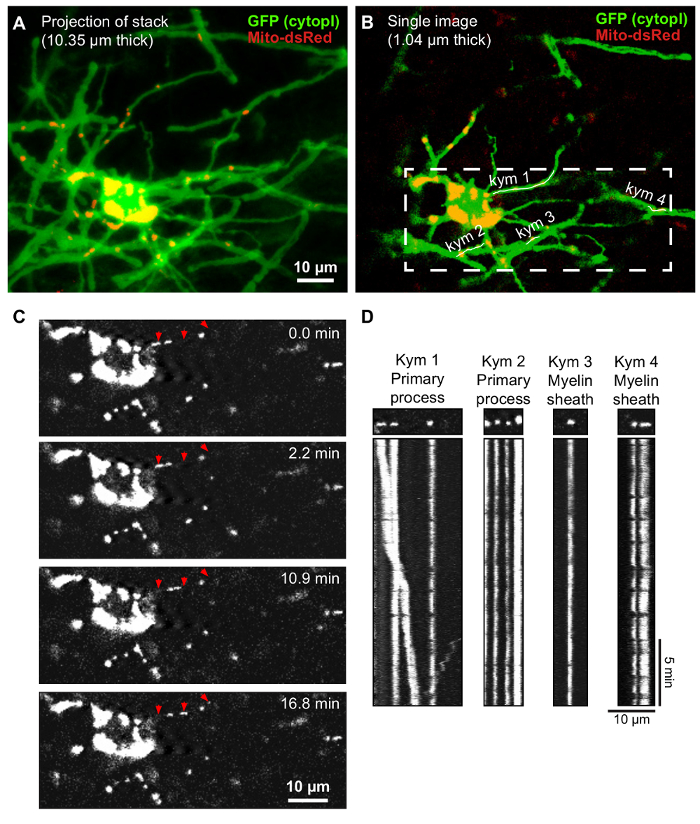
Figure 2: Time-lapse imaging to track mitochondrial movement in oligodendrocytes. (A) Projection of z-stack (optical section 10.35 µm) of oligodendrocyte transduced with MBP_mito_dsred (red mitochondria) and CAG_GFP (green fluorescence). (B) Single confocal image (optical section 1.04 µm) of the same cell. Square indicates region that was selected for time-lapse imaging (C) and the lines drawn to make kymographs (kym) in (D) are indicated. (C) Images from time-lapse recording of the cell in A and B taken at different time points. Moving mitochondria are indicated (red arrows). (D) Kymographs from the trajectories indicated in (B). Stationary mitochondria are seen as straight vertical lines and moving mitochondria are seen as diagonal lines. As can be seen in the kymographs, and in the time-course images in (C), only mitochondria in the primary process marked Kymograph 1 (Kym 1) are moving. The mitochondria in the other three kymographs are stationary during the time-lapse recording. Please click here to view a larger version of this figure.
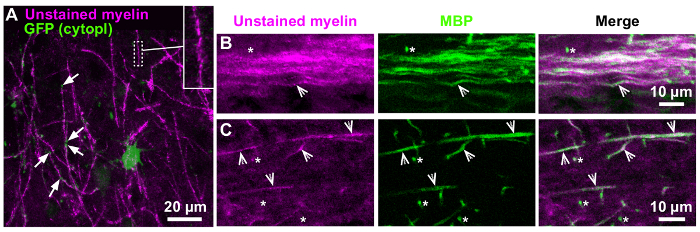
Figure 3: Visualization of unstained myelin by Confocal Reflectance microscopy (CoRe). (A) Projection of z-stack (optical section 10.35 µm) from the neocortex of a live organotypic brain slice. The GFP expression in the oligodendrocyte shown here (green cell body) is too dim for identification of primary processes and myelin sheaths. Instead, myelin sheaths were visualized by excitation at 488 nm and capturing emitted light at 470 – 500 nm (shown in magenta). Some of the cytoplasm-rich paranodes of the GFP+ oligodendrocyte can be seen at the tips of the myelin internodes (arrows). Insert: Magnified part of myelin sheath shows that the myelin visualized with CoRe appears "patchy" or discontinuous (see main text for details). (B-C) Confocal images from the striatum (B) and stratum radiatum of hippocampus CA1 (C) of an acute mouse brain slice that has been fixed in 4% PFA and immunostained for myelin basic protein (MBP). Horisontal myelin sheaths (arrowheads) are visible with CoRe, whereas sheaths that are perpendicular to the field of view (asterisks) are most often not visible. Please click here to view a larger version of this figure.
Reagents' Table: Please click here to download this file.
Discussion
The protocol for making organotypic cultures described here is a modified version18 of the protocol described by De Simoni and Yu (2006)5. The most important changes have been outlined below. Tris buffer is added to the culture medium, which improves the survival of the slices when outside of the incubator during viral transduction and changing of the cell medium. The sterilization procedure for confetti is also changed. While other protocols sterilize confetti by autoclaving, we do not recommend this because it will cause several of the confetti to curl. Avoid using curled confetti as slices grown on these tend to be less healthy. Our protocol uses mice, not rats, and is modified to culture the cortex instead of the hippocampus, which we find works well with thinner slices (230 µm vs 300 µm). As myelination peaks around postnatal day 10 – 20 in mice and rats19, cultures made from mice at p7-9 (and then cultured for 11 – 14 days) are optimal for studying myelinating oligodendrocytes. At 11 – 14 days in vitro, the cultures contain several mature oligodendrocytes with compact myelin sheaths13. Cultures made from younger mice will contain fewer mature oligodendrocytes at the end of the culture period, whereas cultures made from older mice have shown more challenging to keep healthy20.
The procedure for making organotypic slices involves several critical steps. The dissection and mounting of brains, as well as placing slices onto confetti, must be done fast and requires some skill. Typically, the first attempts at making organotypic slice cultures are less successful, but 2-3 rounds of practice should be enough to acquire the skills needed for making healthy cultures. It is important to keep a sterile environment throughout the procedure to avoid contamination of the cultures. Moreover, when working with living cells, it is always imperative to ensure correct pH and osmolality of the solutions to maintain cell health. It is, therefore, a good advice to check osmolality (should be 270-310 mOsm/kg) and pH of all solutions that are used on the slices, especially when doing the experiment for the first time. The culture and dissection solutions for the organotypic cultures contain a pH indicator (phenol red), but the yellow serum in the culture medium can give an impression of a lower than the actual pH. The culture medium contains Penicillin, Streptomycin, and Nystatin to minimize the chances of infection. It is therefore recommended to use these antibiotics and antimycotics when setting up the organotypic slice culture technique in the lab, but they can later be avoided when expert aseptic technique is applied. The current protocol uses cell morphology to assess cell viability. For a more quantitative examination, slices can be loaded with propidium iodide or similar dyes as described elsewhere21. However, it is important to be aware that the emission spectrum of propidium iodide overlaps with dsred, tdtomato and other red indicators. Another limitation of propidium iodide and similar reagents is that they do not readily enter the deeper layers of the slice, thus limiting assessment of cell viability to the top layer of cells5.
A variety of methods exists for gene delivery. The main advantages of using AAV transduction of organotypic slice cultures to visualize organelles are as follows: AAV transduction has a better success rate for postmitotic oligodendrocytes compared with non-viral transfection methods22. Further, in organotypic cultures all brain cell types are present, and oligodendrocytes have a morphology similar to that seen in vivo, including compact myelin13. This is different from oligodendrocytes in monoculture, which lack the communication with axons, and thus do not make compact myelin. Imaging in vitro is easier compared with in vivo and offers a better spatial resolution, which is of particular interest when imaging small organelles such as mitochondria. Future research will aim to image in vivo, but faces a great challenge with the optical aberrations caused by the lipid-rich myelin sheath. Furthermore, AAV serotype tropism and promoter specificity may differ between in vivo and in vitro conditions13,23. In the protocol presented here, AAV2/8 MBP_mito_dsred and AAV2/8 MBP_mito_GFP is used to visualize oligodendrocyte mitochondria. AAV serotype 2/1 was also tested with MBP_mito_dsred but infected a lower number of oligodendrocytes in the organotypic slices (not shown). AAV2/8 CAG_GFP and AAV2/8 CAG_tdtomato was used to visualize oligodendrocyte cytoplasm. Despite using the general CAG promoter for this, AAV 2/8 CAG_GFP and CAG_tdtomato selectively infected oligodendrocytes, with only a small number of astrocytes and neurons being infected (these could be easily identified by their characteristic morphology). This selectivity for oligodendrocytes is presumably due to the tropism of the AAV 2/8 serotype24. However, for organelle-targeted expression, the higher cell-specificity achieved with the MBP- or other oligodendrocyte-specific promoters is recommended. Other serotypes tested with CAG_GFP by us were AAV 2/1, 2/2, 2/5, 2/7, 2/9, most of which infected mainly neurons and astrocytes (not shown).
Depending on the construct and the experimental conditions, modification of the cell transduction will be needed. If the transduced cells appear too dim, try a longer time in vitro after transduction to increase expression. If the cells appear bright but unhealthy, reduce the time after transduction. If very few cells are transduced, the concentration of AAV can be increased. However, it is also possible that the time in vitro after transduction is too long and that transduced cells have died. For the constructs used here, expression levels were optimal 4 – 6 days after transduction. At 7 – 8 days after transduction, cells appeared less healthy, with blobby processes, and 10 days after transduction, only a few transduced cells were visible (not shown).
This protocol uses an upright microscope for the live imaging of organotypic slices. This is different from most mono-and co-cultures that typically use an inverted microscope. It is suboptimal to use an inverted microscope here because the slices must then be turned with the confetti facing upwards, which presumably causes the slices to be less healthy. One advantage of the upright microscope is the possibility to add electrophysiology to the setup. Moreover, the use of a pump to change medium means that drug solutions can easily be added and removed during the time-lapse imaging. A disadvantage with the pump is problems maintaining focus caused by vibrations and currents in the bath. Under the microscope, the cells will gradually become less healthy. We therefore keep slices under the microscope for a maximum of 1 hour. Imaged slices are thrown away (special waste). To ensure the imaging conditions are optimal, imaging of control cells, in which mitochondrial movement is already well characterized, should be run in parallel. For example, mitochondrial movement in axons or dendrites can be imaged by transducing slices with AAV 2/1 Syn_mito_dsred (in which dsred expression is driven by the neuron-specific synapsin promoter) or AAV 2/1 CAG_mito_dsred (followed by immunostaining to confirm cell identity). Analysis of mitochondrial movement can be done using the Multiple kymograph tool in ImageJ as described elsewhere25.
Here we describe CoRe for visualization of unstained myelin during live imaging. Compared with SCoRe9, which uses three excitation wavelengths, CoRe only uses one (488 nm) and is thus simpler to implement. In SCoRe, the three wavelengths each reveal different "pieces" of the myelin sheath and together give a full image of the sheath. In addition, SCoRe can be used to detect myelin sheath structures such as cytoplasmic pockets. With the single wavelength used in CoRe, a less detailed view is given and sheaths may appear granulated or discontinuous (Fig. 3). Nevertheless, CoRe visualizes near 100% of myelin sheaths that are horizontal to the field of view. Thus, CoRe is useful for detection of myelin sheaths, but not for analysis of myelin structures or integrity.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Linda Hildegard Bergersen and Magnar Bjørås for access to cell lab and equipment, Janelia Molecular Biology Shared Resource staff for plasmid and virus production and Koen Vervaeke for assistance with laser power measurements. This work was funded by the Norwegian Health Association, the Norwegian Research Council and the microscopy equipment was funded by Norbrain.
Materials
| Agarose | Sigma | A9539 | |
| BD Microlance 19G | BD | 301500 | Needles used for in- and outlet of bath |
| Bioxide gas | AGA | 105701 | |
| Brand pipette bulbs | Sigma-Aldrich | Z615927 | Pipette bulbs |
| Bunsen burner (Liquid propane burner) | VWR | 89038-530 | |
| Cable assembly for heater controllers | Warner Instruments | 64-0106 | Temperature controller – thermometer part |
| CaCl2 | Fluka | 21100 | |
| CO2 | AGA | 100309 | CO2 for incubator |
| Cover glass, square Corning | Thermo Fischer Scientific | 13206778 | To attach under bath for live imaging. Seal with glue or petrolium jelly. |
| D-(+)-Glucose | Sigma | G7021 | |
| Delicate forceps | Finescience | 11063-07 | For dissection |
| Diamond scriber pen | Ted Pella Inc. | 54463 | |
| Disposable Glass Pasteur Pipettes 230 mm | VWR | 612-1702 | Glass pipettes |
| Double edge stainless steel razor blade | Electron Microscopy Sciences | #7200 | Razor blade for vibratome |
| Earle's Balanced Salt Solution (EBSS) | Gibco-Invitrogen | 24010-043 | |
| Filter paper circles | Schleicher & Schuell | 300,220 | Filter paper used for filtration of PFA |
| Fun tack | Loctite | 1270884 | Use to connect/adjust position of in- and outlets in bath |
| Hand towel C-Fold 2 | Katrin | 344388 | |
| Harp, Flat for RC-41 Chamber, | Warner Instruments | 64-1418 | Harp to hold down confetti in bath. Cut off strings before use with organotypic slices. 1.5 mm, 13mm, SHD-41/15 |
| HEPES, FW: 260.3 | Sigma | H-7006 | |
| Holten LaminAir, Model 1.2 | Heto-Holten | 96004000M | Laminar flow hood |
| Horse serum, heat inactivated | Gibco-Invitrogen | 26050-088 | |
| KCl | Sigma | P9541 | |
| LCR Membrane, PTFE, | Millipore | FHLC0130 | Confetti |
| Leica VT1200 | Leica | 14048142065 | Vibratome |
| MEM-Glutamax with HEPES | Thermo Fischer Scientific | 42360024 | |
| MgCl2 | R.P. Normapur | 25 108.295 | |
| Micro Spoon Heyman Type B | Electron Microscopy Sciences | 62411-B | Small, rounded spatula with sharpened end for dissection |
| Millex-GP filter unit | Millipore | SLGPM33RA | Syringe filter unit |
| Millicell cell culture insert, 30 mm | Millipore | PICM03050 | Cell culture inserts |
| Minipuls 3 Speed Control Module | GILSON | F155001 | Peristaltic pump for live imaging – Control module part (connect to two-cannel head) |
| Na2HPO4 | Sigma-Aldrich | S7907 | |
| NaCl | Sigma-Aldrich | S9888 | |
| NaH2PO4 | Sigma-Aldrich | S8282 | |
| NaHCO3 | Fluka | 71628 | |
| Nunclon Delta Surface | Thermo Fischer Scientific | 140675 | Culture plate |
| Nystatin Suspension | Sigma-Aldrich | N1638 | |
| Objective W "Plan-Apochromat" 40x/1.0 DIC | Zeiss | 441452-9900-000 | Water immersion objective used for live imaging. (WD=2.5mm), VIS-IR |
| Parafilm | VWR | 291-1211 | |
| Paraformaldehyde, granular | Electron Microscopy Sciences | #19208 | |
| PC-R perfusion chamber | SiSkiYou | 15280000E | Bath for live imaging |
| Penicillin-Streptomycin, liquid | Invitrogen | 15070-063 | |
| Petri dish 140 mm | Heger | 1075 | Large Petri dish |
| Petri dish 92×16 mm | Sarstedt | 82.1473 | Medium Petri dish |
| Petridish 55×14,2 mm | VWR | 391-0868 | Small Petri dish |
| Phosphate buffered saline (PBS) | Sigma | P4417 | PBS tablets |
| R2 Two Channel Head | GILSON | F117800 | Peristaltic pump for live imaging – Two channel head part (requires control module) |
| Round/Flat Spatulas, Stainless Steel | VWR | 82027-528 | Large spatula for dissection |
| Sand paper | VWR | MMMA63119 | Optional, for smoothing broken glass pipettes |
| Scissors, 17,5 cm | Finescience | 14130-17 | Large scissors for dissection |
| Scissors, 8,5 | Finescience | 14084-08 | Small, sharp scissors for dissection |
| Single edge, gem blade | Electron Microscopy Sciences | #71972 | Single edge razor blade |
| Single inline solution heater SH-27B | Warner Instruments | 64-0102 | Temperature controller – heater part |
| Steritop-GP Filter unit, 500 ml , 45mm | Millipore | SCGPT05RE | Filter to sterilize solutions |
| Super glue precision | Loctite | 1577386 | |
| Surgical scalpel blade no. 22 | Swann Morton Ltd. | 209 | Rounded scalpel blade |
| Temperature controller TC324B | Warner Instruments | 64-0100 | Temperature controller for live imaging (requires solution heater and cable assembly) |
| Trizma base | Sigma | T1503 | |
| Trizma HCl | Sigma | T3253 | |
| Water jacketed incubator series II | Forma Scientific | 78653-2882 | Incubator |
References
- Barry, C., Pearson, C., Barbarese, E. Morphological organization of oligodendrocyte processes during development in culture and in vivo. Dev. Neursosci. 18, 233-242 (1996).
- Simpson, P. B., Mehotra, S., Lange, G. D., Russell, J. T. High density distribution of endoplasmic reticulum proteins and mitochondria at specialized Ca2+ release sites in oligodendrocyte processes. J. Biol. Chem. 272, 22654-22661 (1997).
- Sterky, F. H., Lee, S., Wibom, R., Olson, L., Larsson, N. G. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc. Natl. Acad. Sci. U. S. A. 1080, 12937-12942 (2011).
- Stoppini, L., Buchs, P. A., Muller, D. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods. 37, 173-182 (1991).
- De Simoni, A., Yu, L. M. Preparation of organotypic hippocampal slice cultures: interface method. Nat. Protoc. 1, 1439-1445 (2006).
- Lim, H., et al. Label-free imaging of Schwann cell myelination by third harmonic generation microscopy. Proc. Natl. Acad. Sci. U. S. A. 111, 18025-18030 (2014).
- Farrar, M. J., Wise, F. W., Fetcho, J. R., Schaffer, C. B. In vivo imaging of myelin in the vertebrate central nervous system using third harmonic generation microscopy. Biophys. J. 100, 1362-1371 (2011).
- Fu, Y., Wang, H., Huff, T. B., Shi, R., Cheng, J. X. Coherent anti-Stokes Raman scattering imaging of myelin degradation reveals a calcium-dependent pathway in lyso-PtdCho-induced demyelination. J. Neurosci. Res. 85, 2870-2881 (2007).
- Schain, A. J., Hill, R. A., Grutzendler, J. Label-free in vivo imaging of myelinated axons in health and disease with spectral confocal reflectance microscopy. Nat. Med. 20, 443-449 (2014).
- Shin, J. H., Yue, Y., Duan, D. Recombinant adeno-associated viral vector production and purification. Methods Mol. Biol. 798, 267-284 (2012).
- Kunkel, T. A. Oligonucleotide-directed mutagenesis without phenotypic selection. Curr. Prot. Neurosci. , 4.10.1-4.10.6 (2001).
- Gow, A., Friedrich, V. L., Lazzarini, R. A. Myelin basic protein gene contains separate enhancers for oligodendrocyte and Schwann cell expression. J. Cell Biol. 119, 605-616 (1992).
- Rinholm, J. E., et al. Movement and structure of mitochondria in oligodendrocytes and their myelin sheaths. Glia. 64, 810-825 (2016).
- Rintoul, G. L., Filiano, A. J., Brocard, J. B., Kress, G. J., Reynolds, I. J. Glutamate decreases mitochondrial size and movement in primary forebrain neurons. J. Neurosci. 23, 7881-7888 (2003).
- Jackson, J. G., O’Donnell, J. C., Takano, H., Coulter, D. A., Robinson, M. B. Neuronal activity and glutamate uptake decrease mitochondrial mobility in astrocytes and position mitochondria near glutamate transporters. J. Neurosci. 34, 1613-1624 (2014).
- Macaskill, A. F., et al. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 61, 541-555 (2009).
- Karadottir, R., Attwell, D. Combining patch-clamping of cells in brain slices with immunocytochemical labeling to define cell type and developmental stage. Nat. Protoc. 1, 1977-1986 (2006).
- Rinholm, J. E., et al. Regulation of oligodendrocyte development and myelination by glucose and lactate. J. Neurosci. 31, 538-548 (2011).
- Davison, A. N., Dobbing, J. Myelination as a vulnerable period in brain development. Br. Med. Bull. 22, 40-44 (1966).
- Humpel, C. Organotypic brain slice cultures: A review. Neuroscience. 305, 86-98 (2015).
- Noraberg, J., Kristensen, B. W., Zimmer, J. Markers for neuronal degeneration in organotypic slice cultures. Brain Res. Protoc. 3, 278-290 (1999).
- Karra, D., Dahm, R. Transfection techniques for neuronal cells. J. Neurosci. 30, 6171-6177 (2010).
- Pignataro, D., et al. Adeno-Associated Viral Vectors Serotype 8 for Cell-Specific Delivery of Therapeutic Genes in the Central Nervous System. Front. Neuroanat. 11 (2), (2017).
- Hutson, T. H., Verhaagen, J., Yanez-Munoz, R. J., Moon, L. D. Corticospinal tract transduction: a comparison of seven adeno-associated viral vector serotypes and a non-integrating lentiviral vector. Gene Ther. 19, 49-60 (2012).
- Neumann, S., Campbell, G. E., Szpankowski, L., Goldstein, L. S. B., Encalada, S. E. Characterizing the composition of molecular motors on moving axonal cargo using cargo mapping analysis. J. Vis. Exp. (92), e52029 (2014).

