Measuring Rates of Herbicide Metabolism in Dicot Weeds with an Excised Leaf Assay
Summary
This manuscript describes how herbicide metabolism rates can be effectively quantified with excised leaves from a dicot weed, thereby reducing variability and removing any possible confounding effects of herbicide uptake or translocation typically observed in whole-plant assays.
Abstract
In order to isolate and accurately determine rates of herbicide metabolism in an obligate-outcrossing dicot weed, waterhemp (Amaranthus tuberculatus), we developed an excised leaf assay combined with a vegetative cloning strategy to normalize herbicide uptake and remove translocation as contributing factors in herbicide-resistant (R) and –sensitive (S) waterhemp populations. Biokinetic analyses of organic pesticides in plants typically include the determination of uptake, translocation (delivery to the target site), metabolic fate, and interactions with the target site. Herbicide metabolism is an important parameter to measure in herbicide-resistant weeds and herbicide-tolerant crops, and is typically accomplished with whole-plant tests using radiolabeled herbicides. However, one difficulty with interpreting biokinetic parameters derived from whole-plant methods is that translocation is often affected by rates of herbicide metabolism, since polar metabolites are usually not mobile within the plant following herbicide detoxification reactions. Advantages of the protocol described in this manuscript include reproducible, accurate, and rapid determination of herbicide degradation rates in R and S populations, a substantial decrease in the amount of radiolabeled herbicide consumed, a large reduction in radiolabeled plant materials requiring further handling and disposal, and the ability to perform radiolabeled herbicide experiments in the lab or growth chamber instead of a greenhouse. As herbicide resistance continues to develop and spread in dicot weed populations worldwide, the excised leaf assay method developed and described herein will provide an invaluable technique for investigating non-target site-based resistance due to enhanced rates of herbicide metabolism and detoxification.
Introduction
Herbicide resistance in weeds presents a serious threat to the global production of food and fiber1,2. Currently, thousands of resistant populations and biotypes from over a hundred weed species worldwide have been documented and studied3. A major mechanism that confers herbicide resistance in plants is the alteration of herbicide target-site genes and proteins, including genetic mutations that affect herbicide-protein binding kinetics or amplification of the target-site gene2. Metabolic detoxification via elevated activities of cytochrome P450 monooxygenase(P450) or glutathione S-transferase (GST) enzymes is another mechanism that confers herbicide resistance in weeds, which is distinct in several ways from target-site-based mechanisms2. Metabolic-based resistance has significant ramifications for whether plant fitness costs (a.k.a. fitness penalties) may result from the herbicide-resistance mechanism, as well as regarding the potential for a single detoxification mechanism to confer cross- or multiple-herbicide resistance in weed populations1,2,4. Generally, herbicide metabolism in plants can be divided into three distinct phases5. Phase I involves herbicide conversion or activation such as P450-mediated hydroxylation of aromatic rings or alkyl groups, or by N– or O-dealkylation reactions, leading to increased polarity and partial herbicide detoxification5,6. Newly introduced functional groups in Phase I can provide linkage sites for conjugation to reduced glutathione by GSTs or to glucose by UDP-dependent glycosyltransferases in Phase II5,7. For example, the major initial metabolite of primisulfuron-methyl in maize is hydroxy-primisulfuron-methyl8, which can be further metabolized to hydroxy-primisulfuron-glucoside (Phase II) and then transported to the vacuole for long-term storage or further metabolic processing5,6 (Phase III).
Waterhemp (Amaranthus tuberculatus) is a difficult-to-control, dicot annual weed species that hinders the production of maize (Zea mays), soybean (Glycine max), and cotton (Gossypium hirsutum) in the United States. The high degree of genetic diversity of waterhemp is facilitated by its dioecious biology and long-distance wind pollination, and a single female waterhemp plant can produce up to a million seeds9. These seeds are small and easily spread, which naturally endow waterhemp with an effective dispersal mechanism. Waterhemp displays continuous germination throughout the growing season9, and its seeds are able to germinate after several years of dormancy. Waterhemp is a C4 plant that possesses a higher growth rate than most broadleaf weeds in arable cropping systems10. In addition, numerous waterhemp populations are resistant to multiple families of herbicides3.
A population of waterhemp (designated MCR) from Illinois is resistant to 4-hydroxy-phenylpyruvate dioxygenase (HPPD)-inhibiting herbicides11, such as mesotrione, as well as to atrazine and acetolactate synthase (ALS)-inhibiting herbicides, including primisulfuron-methyl, due to non-target-site based mechanisms12,13. A different population of waterhemp designated ACR14, which is primisulfuron-methyl-resistant (due to a mutation in the ALS gene) and atrazine-resistant but sensitive to mesotrione, and a waterhemp population designated WCS14 that is sensitive to primisulfuron-methyl, mesotrione, and atrazine were used in comparison with MCR in our prior research12 and current experiments (summarized in Table 1). Initial studies did not detect alterations in the HPPD gene sequence or expression levels, or reduced mesotrione uptake, in the MCR population when compared with mesotrione-sensitive populations12. However, metabolism studies with whole plants demonstrated significantly lower levels of parent mesotrione herbicide in MCR compared with ACR and WCS, which correlated with previous phenotypic responses to mesotrione11,12.
| Waterhemp Population | Abbreviation | Phenotype to Mesotrione | Mesotrione Resistance Mechanism | Phenotype to Primisulfuron | Primisulfuron Resistance Mechanism |
| McLean County-Resistant | MCR | Resistant | Metabolism* | Resistant | Metabolism |
| Adams County-Resistant | ACR | Sensitive | – | Resistant | Target-site mutation in ALS14 |
| Wayne County-Sensitive | WCS | Sensitive | – | Sensitive | – |
* Non-target-site resistance mechanisms, other than enhanced metabolism, may also confer mesotrione resistance in the MCR population12.
Table 1: Description of waterhemp populations from Illinois used in this study.
In addition to determining rates of herbicide metabolism in intact waterhemp seedlings, a different experimental approach was developed and employed in our previous research to investigate metabolism by using an excised waterhemp leaf assay12 as well as various P450 inhibitors (e.g., tetcyclacis and malathion). This method was adapted specifically for waterhemp from a previous investigation of primisulfuron-methyl metabolism in excised maize leaves15, since the excised leaf assay had not yet been reported for conducting herbicide metabolism research in a dicot plant. The organophophosate insecticide malathion has been frequently used for in vivo and in vitro herbicide-metabolism research to indicate P450 involvement16. For example, tolerance and rapid metabolism of mesotrione in maize are due to P450-catalyzed ring hydroxylation, which was verified when malathion increased maize sensitivity to mesotrione17. Similarly, malathion inhibited metabolism of the ALS inhibitor primisulfuron-methyl in excised maize leaves15. A major advantage of the excised leaf technique is that data generated are independent of whole-plant translocation patterns, an important factor to consider when assessing metabolism of systemic, postemergence herbicides in plants. Consequently, this method allows quantitative and qualitative metabolic analyses to focus on a single treated leaf12.
A vegetative cloning strategy, in combination with the excised leaf protocol, was previously utilized in waterhemp to conduct metabolism studies12. Due to the outcrossing nature of waterhemp (separate male and female plants), and large degree of genetic diversity within dioecious Amaranthus species9, this protocol ensured that genetically-identical waterhemp seedlings were analyzed within the time-course experiments. This article demonstrates the utility of the excised leaf method for measuring rates of herbicide metabolism in a dicot weed (waterhemp). The quantity of parent herbicide remaining was determined at each time point (Figure 1) by non-linear least squares regression analysis, and was fit with a simple first-order curve in order to estimate the time for 50% of absorbed herbicide to degrade (DT50). Representative chromatograms from reversed-phase high performance liquid chromatography (RP-HPLC) are displayed for ALS-resistant and -sensitive waterhemp populations, which indicate the disappearance of parent herbicide and concomitant formation of polar metabolite(s) during a time-course study (Figure 2). The focus of our article is to describe and demonstrate the utility of the excised leaf assay in combination with a vegetative cloning method for determining precise and reproducible rates of herbicide metabolism in dicot plants, using uniformly ring-labeled (URL-14C) herbicides in three waterhemp populations that differ in their whole-plant responses to HPPD- and ALS-inhibiting herbicides (Table 1).
Protocol
1. Plant Material, Growth Conditions, and Vegetative Cloning
Note: Three waterhemp populations were investigated in this research: MCR (from McLean County, IL), ACR (from Adams County, IL), and WCS (from Wayne County, IL) ( Table 1).
- Collect and suspend waterhemp seeds in 0.1 g L-1 agar:water solution at 4 °C for at least 30 days to enhance germination. Note: Some waterhemp populations are dormant, but this step helps to overcome dormancy and make seed germinate more uniformly.
- Germinate seeds of each waterhemp population (as in Table 1) in 12 x 12 cm trays containing a potting medium in the greenhouse.
- Maintain greenhouse conditions at 28/22 °C day/night temperatures within a 16/8 hr photoperiod12.
- Supplement natural light in the greenhouse with light from mercury halide lamps to provide a minimum of 500 µmol m-2 sec-1 photon flux at the plant canopy level.
- Transplant emerged seedlings when they are 2 cm tall into 80 cm3 pots in the greenhouse.
- Transplant seedlings into 950 cm3 pots containing a 3:1:1:1 mixture of potting mix:soil:peat:sand when seedlings are 4 cm tall. Add 5 g of slow-release fertilizer granules to each pot and mix with this soil mixture.
- Cut and remove the shoot apical meristem containing the three youngest leaves from 7 cm tall plants to eliminate apical dominance.
Note: This step ensures that each plant will grow enough 3 cm axillary shoots to clone. - Excise five axillary shoots (3 cm in length) when seedlings are about 14 cm tall from each waterhemp population (as in Table 1). Remove the majority of the leaves on these axillary shoots to decrease water loss upon further handling, but retain the two youngest leaves to allow further growth upon transplanting.
Note: Handle carefully and try not to damage the shoots when removing the leaves. Label each plant with a stake. - Transplant axillary shoots into 80 cm3 pots (one seedling per pot) into potting medium, which is the same medium used for germination. Potting medium should be saturated initially until roots form in the growth chamber, then maintain constant moisture and place pots in the growth chamber for 7 days to establish roots.
Note: Plants are very sensitive at this point and should not be stressed.- Maintain growth chamber conditions at 28/22 °C day/night temperatures within a 16/8 hr photoperiod12.
- A combination of incandescent and fluorescent bulbs can be used to provide light in the growth chamber and should deliver at least 550 µmol m-2 sec-1 photon flux at plant canopy level12.
- Transplant again into 950 cm3 pots (one seedling per pot) containing a 3:1:1:1 mixture of potting mix:soil:peat:sand (also include 5 g slow-release fertilizer granules) when cloned plants are 4 cm tall, then transfer back to the greenhouse.
- Clone the plants a second time by using the same method described above (i.e., repeat steps from 1.5 to 1.8, and then proceed to step 2.1). For a typical time-course study, generate clones from at least six independent plants from each waterhemp population and record them as lines MCR1-6, ACR1-6 and WCS1-6.
- Establish at least six distinct “parent” clonal lines in order to repeat the experiment and adequately represent genetic variability within each waterhemp population.
- Depending on how many time points will be analyzed, generate enough sub-clones from each “parent” clonal line to conduct a time-course study.
2. Conducting a Metabolism Study with Excised Waterhemp Leaves
- Excise the third-youngest leaves (2 to 3 cm in length; include 0.5 cm of attached petiole) when cloned waterhemp plants are 10 to 12 cm tall for herbicide metabolism and DT50 analysis.
- Cut leaf petioles from each waterhemp plant a second time (ensuring that 0.3 cm of the petiole remains) under water, and place these cut ends into 1.5 ml plastic vials (one leaf per vial)15.
Note: Cutting a second time under water prevents air bubbles from forming and plugging xylem vessels.- Firstly, partially immerse excised leaves with cut petiole in pre-incubation buffer (200 µl) of 0.1 M Tris-HCl (pH 6) for 1 hr.
- Secondly, move each leaf to a new 1.5 ml vial containing an incubation buffer (200 µl) of 0.1 M Tris-HCl (pH 6) plus 150 µM [URL-14C] herbicide, and incubate at 28 °C for 1 hr to allow for herbicide absorption into the leaves.
Note: Both 14C-mesotrione and 14C-primisulfuron-methyl were used in our studies at specific activities of about 20 mCi mmol-1. Both diffusion and uptake through the xylem transpiration stream contribute to the amount of herbicide taken up per leaf.
Note: The effect of metabolic inhibitors on herbicide metabolism can be determined by modifying steps 2.2.1 and 2.2.2 above. Add 100 µM of the inhibitor to the pre-incubation buffer and let incubate for 2 hr, then transfer to incubation buffer with herbicide and again add 100 µM inhibitor. For example, malathion and tetcyclacis inhibit certain cytochrome P450 activities in plants15-17. - Wash the immersed portion of excised leaves with deionized water and transfer excised leaves to a quarter-strength Murashige and Skoog (MS) salts solution (500 µl) for 0 hr, 5 hr, 11 hr, 23 hr, or 35 hr, respectively, in the growth chamber for the time-course metabolism study.
- Harvest by removing leaves from incubation solution at the appropriate time point and manually grind each leaf tissue in liquid nitrogen using a polypropylene tube (15 ml) and glass pestle.
- Extract radiolabeled herbicide and its metabolites from each leaf with 7 ml of 90% acetone with a laboratory tissue homogenizer in the same polypropylene tube as above. Rinse tissue homogenizer with an additional 7 ml of 90% acetone, and continue to homogenize until tissue is thoroughly pulverized (usually about 2 min). Note: 90% acetone can be used for the extraction of most herbicides from plant tissues. However, if this method is unable to extract more than 90% of the radiolabeled compounds absorbed, then 90% methanol or 90% acetonitrile can be substituted as the extraction solvent.
- Label 90% acetone extracts (14 ml total) and store at -4 °C for 16 hr to allow for additional extraction to occur.
Note: Following this extraction of treated leaves, the amount of extractable radioactivity is typically at least 98% of radiolabeled compounds absorbed by the treated leaf. Non-extractable radioactivity (bound residues) in excised leaves averaged only 0.3% across all time points in our previous study12, but this amount can vary according to the herbicide used and time point examined. - Centrifuge samples at 5,000 x g for 10 min and concentrate supernatants at 40 °C with a rotary evaporator until a final volume of 0.5 ml is obtained. Transfer this volume to a 2.0 ml plastic tube.
- Add acetonitrile:water (1:1, by volume) to adjust the final volume of the plant extracts to 1.25 ml and re-centrifuge at 10,000 x g for 10 min to remove any particulates.
- Measure total radioactivity in an aliquot taken from each sample (dpm µl-1) by liquid scintillation spectroscopy (LSS)12 and normalize amounts of [14C-labeled] compounds injected for HPLC analysis (for example, 10,000 total dpm/sample/run) so y-axes will be uniform among samples when graphing results.
3. HPLC Analysis of Herbicide Metabolism in Excised Leaves
Note: Resolve total extractable radioactivity into parent herbicide and herbicide metabolites using the following RP-HPLC conditions12.
- Perform RP-HPLC with a C18 column (4.6 x 250 mm, 5 µm particle size for analytical HPLC) at a flow rate of 1 ml min-1 for most non-polar herbicides.
Note: C4 or C12 RP-HPLC columns can also be used to optimize separation of the parent herbicide from its metabolites. A pre-column filter and/or guard column may also be used to protect and extend the life of the RP column.- Use the following mobile phase for RP-HPLC: Eluent A, 0.1% formic acid (v/v) in water, and Eluent B 100% HPLC-grade acetonitrile.
Note: HPLC-grade methanol can also be used as Eluent B for resolving more polar compounds and metabolites, but be aware that retention times may also change.
Note: The elution profile used to resolve mesotrione or primisulfuron-methyl from their polar metabolites in our research 12 is: Step 1, A:B (4:1, v/v) to A:B (3:2, v/v) in a linear gradient (12 min); Step 2, A:B (3:2, v/v) to A:B (3:7, v/v) in a linear gradient (5 min); Step 3, A:B (3:7, v/v) to A:B (1:9, v/v) in a linear gradient for 2 min (comprising a total of 19 min).
Note: Gradients and isocratic steps may need to be adjusted based on the physical properties of the parent material in order to optimize resolution. - Follow the above linear gradient steps with A:B (1:9, v/v) to A:B (4:1, v/v) in a linear gradient (3 min) and A:B (4:1, v/v) isocratic step (for at least 2 min) to re-equilibrate the RP column before injecting the next extract12.
- Use the following mobile phase for RP-HPLC: Eluent A, 0.1% formic acid (v/v) in water, and Eluent B 100% HPLC-grade acetonitrile.
- Detect and visualize radiolabeled compounds with a Flow Scintillation Analyzer. Record the amount of parent herbicide remaining in each sample as a percentage of total radioactivity in each sample (detected and quantified by the Flow Scintillation Analyzer according to the manufacturer’s instructions) to determine rates of herbicide metabolism in each waterhemp population during the time course.
4. Statistical Procedures
- Arrange treatments in a completely randomized design (or other suitable arrangement for statistical analysis).
- Perform two independent experiments with each treatment, comprised of three biological replicates for each experiment. The two independent experiments include six biological replicates in total. If the experiment effect is non-significant (α = 0.05), combine and analyze data from each independent experiment. If the experiment effect is significant, then analyze each experiment separately.
Note: Include the first three biological replicates in Experiment 1 and the next three biological replicates in Experiment 2. Each biological replicate represents a distinct “parent” clonal line derived from each population (MCR1-6, ACR1-6, and WCS1-6) so that each time-course analysis utilizes genetically-identical plants. The two independent experiments include six biological replicates in total, which allows for adequate representation of genetic variability and determination of a median DT50 for each waterhemp population. - Analyze the data with non-linear least squares regression analysis and fit them with a simple first-order curve in order to estimate DT50s. The equation below describes the model:
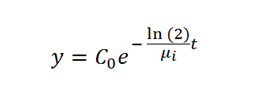
- where in this model y represents the percentage of the parent herbicide remaining at time t, µi is the DT50 for each waterhemp population i, and the parameter C0 is the estimated amount of parent herbicide present at t = 012.
Representative Results
Large differences in rates of mesotrione metabolism were detected between either WCS or ACR and MCR (Figure 1). At each time point, MCR had metabolized mesotrione more rapidly than the two mesotrione-sensitive populations, WCS and ACR, which correlates with previous whole-plant phenotypic responses11. By cloning enough plants from a single parental plant from each population, herbicide metabolism time-course analyses are uniform and reproducible due to the lack of genetic variability within each time course12. For example, the time course for mesotrione metabolism in clonal line 512 is displayed in Figure 1. A total of six different clonal lines, derived from six individual parent plants, were analyzed from each population (Table 1) to determine an overall median DT50 value, which indicated that mesotrione metabolism in MCR was significantly faster (i.e., had a shorter DT50 value) than ACR or WCS12. Additionally, our previous study with excised leaves showed that P450 inhibitors significantly decreased mesotrione metabolism in MCR and maize, but not in ACR, and the major initial metabolite detected in MCR was identified as 4-hydroxy-mesotrione, indicating that enhanced rates of P450-mediated metabolism are associated with resistance in MCR12.
In addition to studying the mechanisms for mesotrione resistance in MCR, a related objective was to determine the mechanism for resistance to ALS inhibitors in MCR. Prior research demonstrated that a sub-population derived from MCR was ALS resistant but did not possess a mutation in the ALS target site gene, indicating a non-target-site based mechanism13. This is in contrast with ACR, which is ALS resistant due to a target site mutation that confers a less-sensitive ALS enzyme14. In order to test this hypothesis, the excised leaf assay was conducted with [URL-14C] primisulfuron-methyl, an ALS-inhibiting herbicide of the sulfonylurea family. The peak area of 14C-primisulfuron-methyl in a representative HPLC chromatogram (retention time of about 21.6 min) was significantly smaller in MCR than in ACR and WCS at 12 HAT (Figure 2). Additionally, two polar metabolites with shorter retention times than primisulfuron-methyl were detected in excised leaf extracts from all three populations (Figure 2). Although the structures of these two polar metabolites were not determined in this study, their rapid formation is consistent with ring hydroxylation of primisulfuron-methyl8 (Phase I metabolism) followed by glucose conjugation5 (Phase II metabolism).
In combination with the qualitative analyses of primisulfuron metabolism in MCR, ACR, and WCS (Figure 2), the amount of parent herbicide remaining at each time point was quantified and used to determine the DT50 values for primisulfuron-methyl in MCR (13.6 hr), ACR (>24 hr), and WCS (>24 hr) (Figure 3). The significantly shorter DT50 value in MCR is consistent with increased metabolism as the major mechanism for ALS resistance in this population, in accord with previous whole-plant research13. Interestingly, however, the DT50 values for primisulfuron in ACR and WCS are similar yet significantly longer than in MCR (Figure 3), despite both MCR and ACR displaying ALS-inhibitor resistant phenotypes (Table 1). The relatively long DT50 in ACR is possibly because it is not necessary to rapidly metabolize primisulfuron, since ACR has a less-sensitive ALS enzyme as its main resistance mechanism14. In summary, the excised waterhemp leaf assay utilized in our research yielded valuable qualitative and quantitative data and provided new insight into metabolism-based resistance mechanisms towards two different herbicides in MCR, ACR, and WCS.
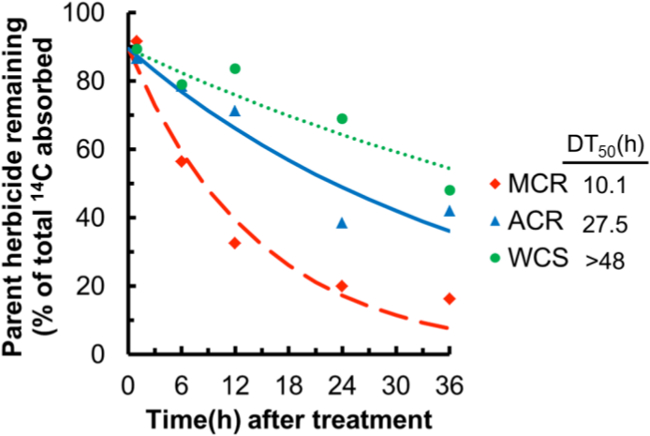
Figure 1: Time course of mesotrione metabolism in excised waterhemp leaves from vegetatively cloned populations. Excised leaves (third-youngest leaf; 2 to 3 cm in length) from waterhemp seedlings (10 to 12 cm) were vegetatively cloned so that each time-course study for each line consisted of genetically-identical plants. Excised leaves were placed in 0.1 M Tris-HCl buffer (pH 6) for 1 hr, followed by 0.1 M Tris-HCl (pH 6) plus 150 µM radiolabeled mesotrione for 1 hr (‘pulse’), then quarter-strength Murashige and Skoog (MS) salts solution (‘chase’) for 0 hr, 5 hr, 11 hr, 23 hr, and 35 hr to allow metabolism to occur. Data were analyzed by non-linear least squares regression analysis and fit with a simple first-order curve to estimate a DT50 separately for each cloned waterhemp population12.The MCR clonal line rapidly metabolized mesotrione, while the ACR and WCS clonal lines metabolized mesotrione more slowly. Data shown are time courses from only clonal line 5 for each waterhemp population, which has been modified from our previous paper12 (Copyright by the American Society of Plant Biologists).
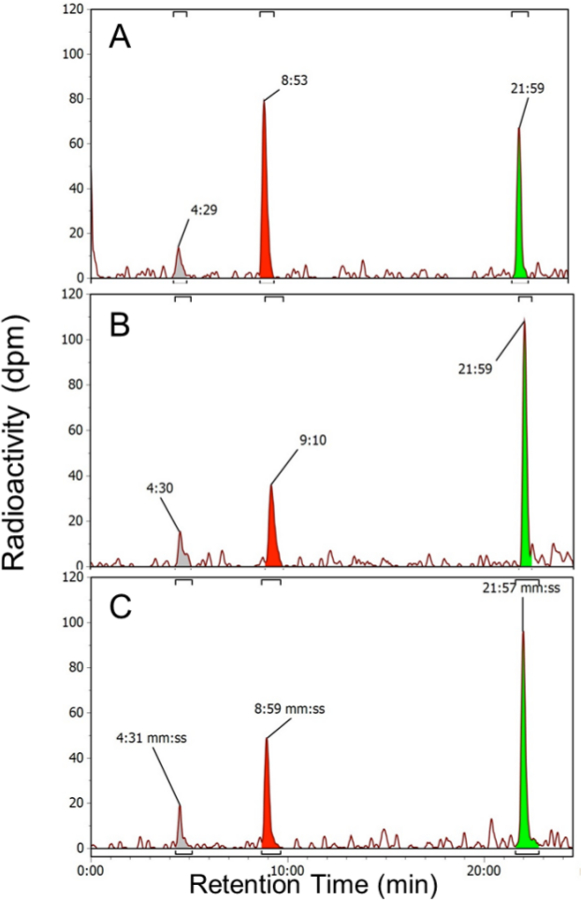
Figure 2: Primisulfuron-methyl metabolism in MCR, ACR, and WCS (12 HAT). Representative reversed-phase HPLC chromatograms for excised waterhemp leaf extracts (12 HAT) supplied with 150 µM radiolabeled primisulfuron-methyl (as described in the Protocol) from McLean County (MCR, A), Adams County (ACR, B), and Wayne County herbicide-sensitive (WCS, C) populations (Table 1). Radioactive peak with a retention time of 21.6 min contains primisulfuron-methyl, as determined by comparison with an authentic analytical standard. Peaks with shorter retention times are likely polar, less phytotoxic metabolites of primisulfuron-methyl, such as hydroxy-primisulfuron-methyl8 or glycosylated forms of hydroxylated metabolites5,6. The MCR population displayed a significant decrease in the amount of parent primisulfuron-methyl relative to ACR and WCS, which is quantified as a lower DT50 for MCR (Figure 3).
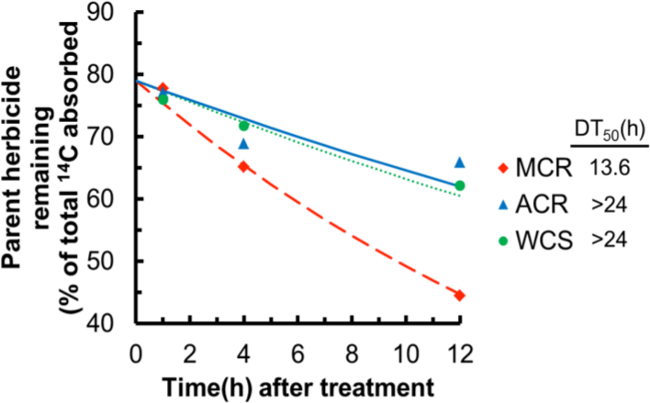
Figure 3: Time course of primisulfuron-methyl metabolism in excised waterhemp leaves. Excised waterhemp leaves (third-youngest leaf; 2 to 3 cm) were treated as described in Figure 1 and our previous research with mesotrione12, except that 150 µM radiolabeled primisulfuron was included as the ‘pulse’ and leaves were incubated in the MS salts solution (‘chase’) for 0 hr, 3 hr, and 11 hr. Data were analyzed by non-linear least squares regression analysis as described previously for mesotrione metabolism (Figure 1)12. The MCR population rapidly metabolized primisulfuron-methyl (DT50 = 13.6 hr), while ACR and WCS metabolized primisulfuron-methyl more slowly (DT50 >24 hr).
Discussion
The excised-leaf method described herein has been used previously in researching primisulfuron metabolism in maize leaves15, but our results demonstrate that this protocol is also effective, accurate, and reproducible for measuring herbicide metabolism in a dicot weed species12. A major advantage of the excised leaf technique compared with whole-plant studies is that an excised leaf is independent of whole-plant translocation patterns of postemergence, systemic herbicides or differences in herbicide uptake among plants or populations. In addition, environmental variability is reduced since the excised leaf assays are conducted in a growth chamber, with a single leaf placed in a tube, compared with studying whole plants grown under greenhouse conditions. A vegetative cloning strategy was also included in our method for studying mesotrione-resistance mechanisms in MCR12 to minimize the large degree of genetic variance within and between Amaranthus populations9. Genetic diversity within weedy Amaranthus populations is illustrated by the amount of variability documented in whole-plant responses to several families of postemergence herbicides18. When conducting time-course studies to determine accurate DT50s and for detecting significant differences when comparing DT50 values between waterhemp populations12, these steps (as outlined in our excised leaf and vegetative cloning protocol) are critical for eliminating or reducing genetic and environmental variability.
Different mechanisms exist in plants for conferring herbicide resistance1,2. For example, waterhemp resistance to ALS-inhibiting herbicides can be target-site based14 or non-target-site based13. Metabolic rates of primisulfuron-methyl (Figure 3) clearly show these differences between two ALS-resistant waterhemp populations, MCR and ACR (Table 1). In combination with PCR amplification and sequence analysis of herbicide target-site genes, the excised leaf assay will greatly assist towards identifying whether herbicide resistance in waterhemp or other dicot weeds is conferred by target site or non-target-site mechanisms2.
Limitations of our excised leaf method include the inability to visualize or measure herbicide translocation patterns throughout the entire plant, or determine if cellular or sub-cellular sequestration mechanisms exist that confer non-target-site based resistance in weeds2. As mentioned previously, this aspect was also considered an advantage when determining precise herbicide metabolism rates in mesotrione-resistant weed populations12, but conversely could be considered a drawback when studying systemic herbicides that are not metabolized significantly in plants (i.e., non-selective herbicides) such as glyphosate2, or contact herbicides that are non-selective in natural weed populations, such as paraquat or glufosinate19. However, if target-site mechanisms are not revealed initially then enhanced rates of herbicide metabolism are typically investigated next1,4, particularly when studying weed resistance to HPPD-inhibiting herbicides such as mesotrione12, ALS-inhibiting herbicides such as primisulfuron-methyl13, photosystem II-inhibiting herbicides such as atrazine1,12, or acetyl-CoA-inhibiting herbicides4.
P450s have been associated with ALS resistance in an Echinochloa population20, a weedy grass species, as well as with mesotrione and ALS resistance in MCR12,13. A dioecious, dicot weed related to waterhemp, Palmer amaranth (A. palmeri), is also prone to developing herbicide resistance via several different mechanisms1-3. As more herbicide-resistant weed populations and species are documented throughout the world3, the need for rapidly examining herbicide metabolism as a potential resistance mechanism will continue to increase. The excised leaf approach, which is distinct but related to whole-plant studies typically used to investigate herbicide resistance mechanisms, is an accurate and valuable tool to evaluate and quantify herbicide metabolism in plants. As a result, the capability to perform these analyses with accurate, reproducible methods including the excised leaf assay will greatly assist researchers in determining non-target-site resistance mechanisms in both grass4 and dicot12 weeds.
Future applications of this method may also include determining rates of herbicide metabolism in dicot crops such as soybean and cotton. Ongoing research in our laboratory with herbicide-tolerant soybean varieties has also utilized the excised leaf approach. However, since soybeans are a legume crop with trifoliate leaves, an ‘excised petiole’ technique has been adapted and developed so that each trifoliolate leaf can absorb radiolabeled herbicide via uptake through the main petiole (Skelton, Lygin, and Riechers, unpublished data).
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thank Wendy Zhang, Austin Tom, Jacquie Janney, Erin Lemley, and Brittany Janney for assistance with plant growth and extractions, Dr. Anatoli Lygin for assistance with chromatographic analyses, and Syngenta Crop Protection for funding.
Materials
| Agar | Sigma-Aldrich | A1296 | for pre-germinating seeds |
| Potting medium | Sun Gro Horticulture | 49040233 | for plant growth |
| Nutricote | Agrivert | TOTAL BLEND 13-13-13 T100 | slow-release fertilizer |
| Growth chamber E15 | Controlled Environments Limited | 20207 | plant culturing |
| Tris base | Fisher Scientific | BP152-500 | buffer for excised leaves |
| HCl (concentrated) | Fisher Scientific | A144500 | adjust pH of buffer |
| Murashige and Skoog (MS) salts | Sigma-Aldrich | M0404 | incubation of excised leaves |
| Methanol | Fisher Scientific | A452-4 | leaf washes after incubation |
| Acetone | Sigma-Aldrich | 179124 | plant extractions |
| Acetonitrile (HPLC grade) | Macron Fine Chemicals | MKH07610 | HPLC mobile phase |
| Formic acid | Mallinckrodt Analytical | MK259205 | acidify mobile phase pH |
| Micro-centrifuge | Eppendorf | 5417R | 1.5 or 2.0 mL tubes |
| Centrifuge (temperature controlled) | Eppendorf | 5810R | 15 or 50 mL tubes |
| Polypropylene centrifuge tube | Corning Inc. | 430790 | 15 mL, sterile |
| Rotary evaporator | BÜCHI | R200 | concentrate plant samples |
| Liquid scintillation spectrometry (LSS) | Packard Instruments | 104470 | quantify 14C |
| High-performance liquid chromatography | Perkin Elmer | N2910401 | resolve herbicide metabolites |
| Flow scintillation analyzer | LabLogic System | 1103303 | for HPLC analysis of 14C |
| Hypersil Gold C18 column | Thermo-Scientific | 03-050-522 | reversed phase |
| Ultima-Flo M cocktail | Perkin Elmer | 6013579 | for Flow-scintillation analyzer |
| Scintillation Cocktail (ScintiVerse BD) | Fisher Scientific | SX18 | for LSS; biodegradable |
| Laboratory homogenizer | Kinematica | CH-6010 | homogenize leaf samples |
Riferimenti
- Yu, Q., Powles, S. Metabolism-based herbicide resistance and cross-resistance in crop weeds: A threat to herbicide sustainability and global crop production. Plant Physiology. 166, 1106-1118 (2014).
- Powles, S. B., Yu, Q. Evolution in action: plants resistant to herbicides. Annual Reviews in Plant Biology. 61, 317-347 (2010).
- Heap, I., et al. Global perspective of herbicide-resistant weeds. Pest Management Science. 70 (9), 1306-1315 (2014).
- Délye, C., et al. Non-target-site-based resistance should be the centre of attention for herbicide resistance research: Alopecurus myosuroides as an illustration. Weed Research. 51 (5), 433-437 (2011).
- Kreuz, K., Tommasini, R., Martinoia, E. Old enzymes for a new job. Herbicide detoxification in plants. Plant Physiology. 111, 349-353 (1996).
- Riechers, D. E., Kreuz, K., Zhang, Q. Detoxification without intoxication: herbicide safeners activate plant defense gene expression. Plant Physiology. 153, 3-13 (2010).
- Siminszky, B. Plant cytochrome P450-mediated herbicide metabolism. Phytochemistry Reviews. 5 (2-3), 445-458 (2006).
- Fonné-Pfister, R., et al. Hydroxylation of primisulfuron by an inducible cytochrome P450-dependent monooxygenase system from maize. Pesticide Biochemistry and Physiology. 37 (2), 165-173 (1990).
- Steckel, L. E. The dioecious Amaranthus spp.: here to stay. Weed Technology. 21 (2), 567-570 (2007).
- Horak, M. J., Loughin, T. M. Growth analysis of four Amaranthus species. Weed Science. 48 (3), 347-355 (2000).
- Hausman, N. E., et al. Resistance to HPPD-inhibiting herbicides in a population of waterhemp (Amaranthus tuberculatus) from Illinois, United States. Pest Management Science. 67 (3), 258-261 (2011).
- Ma, R., et al. Distinct detoxification mechanisms confer resistance to mesotrione and atrazine in a population of waterhemp. Plant Physiology. 163, 363-377 (2013).
- Guo, J., et al. Non-target-site resistance to ALS inhibitors in waterhemp (Amaranthus tuberculatus). Weed Science. in press, (2015).
- Patzoldt, W. L., Tranel, P. J., Hager, A. G. A waterhemp (Amaranthus tuberculatus) biotype with multiple resistance across three herbicide sites of action. Weed Science. 53 (1), 30-36 (2005).
- Kreuz, K., Fonné-Pfister, R. Herbicide-insecticide interaction in maize: malathion inhibits cytochrome P450-dependent primisulfuron metabolism. Pesticide Biochemistry and Physiology. 43 (3), 232-240 (1992).
- Correia, M. A., Ortiz de Montellano, P. R., Ortiz de Montellano, P. R. . Cytochrome P450: Structure, Mechanism, and Biochemistry. , 247-322 (2005).
- Hawkes, T. R., et al. Mesotrione: mechanism of herbicidal activity and selectivity in corn. Proceedings of the Brighton Crop Protection Conference – Weeds. 2, 563-568 (2001).
- Patzoldt, W. L., Tranel, P. J., Hager, A. G. Variable herbicide responses among Illinois waterhemp (Amaranthus rudis and A. tuberculatus) populations. Crop Protection. 21 (9), 707-712 (2002).
- Jalaludin, A., Yu, Q., Powles, S. B. Multiple resistance across glufosinate, glyphosate, paraquat and ACCase-inhibiting herbicides in an Eleusine indica population. Weed Research. 55 (1), 82-89 (2015).
- Iwakami, S., et al. Cytochrome P450 CYP81A12 and CYP81A21 are associated with resistance to two acetolactate synthase inhibitors in Echinochloa phyllopogon. Plant Physiology. 165, 618-629 (2014).

