模拟信号蛋白的功能:走向人工信号转导疗法
Summary
We present guidelines for developing synthetic ‘chemical transducers’ that can induce communication between naturally unrelated proteins. In addition, detailed protocols are presented for synthesizing and testing a specific ‘transducer’ that enables a growth factor to activate a detoxifying enzyme and consequently, to regulate the cleavage of an anticancer prodrug.
Abstract
Signal transduction pathways, which control the response of cells to various environmental signals, are mediated by the function of signaling proteins that interact with each other and activate one other with high specificity. Synthetic agents that mimic the function of these proteins might therefore be used to generate unnatural signal transduction steps and consequently, alter the cell’s function. We present guidelines for designing ‘chemical transducers’ that can induce artificial communication between native proteins. In addition, we present detailed protocols for synthesizing and testing a specific ‘transducer’, which can induce communication between two unrelated proteins: platelet-derived growth-factor (PDGF) and glutathione-S-transferase (GST). The way by which this unnatural PDGF-GST communication could be used to control the cleavage of an anticancer prodrug is also presented, indicating the potential for using such systems in ‘artificial signal transduction therapy’. This work is intended to facilitate developing additional ‘transducers’ of this class, which may be used to mediate intracellular protein-protein communication and consequently, to induce artificial cell signaling pathways.
Introduction
信号转导途径在几乎所有的细胞过程中发挥显著作用,并允许细胞迅速环境信号作出反应。1这些途径通常由结合的信号传导分子的触发以胞外受体,这导致细胞内酶的活化。扩增和细胞内的该信号的传播受信令形成蛋白质 – 蛋白质相互作用的网络,其中酶可逆地以高特异性活性的蛋白质的功能介导的。因为这些网络的失调经常导致癌症的发展,出现了在建立“的癌症的信号转导的治疗',2由此药物被设计为破坏恶性信号通路的兴趣。我们最近已提出了另一种方法的信号,依赖于药物来产生不自然的信号转导途径的能力转导的治疗。 <suP> 3特别是,我们相信,通过设计模拟信号蛋白的功能的合成剂,这将有可能间接地调节细胞的功能。例如,这些人工网络可以使蛋白质生物标志物来激活切割前体药物的酶。或者,这些信号蛋白模拟物可能能够激活非天然细胞信号传导途径,导致治疗效果。
为了证明这种方法的可行性,我们最近创建的合成“化学换能器'4,使血小板衍生的生长因子(PDGF)通过激活谷胱甘肽-S-转移酶(GST),这是引发抗癌前药的切割不是其天然结合伴侣。这个'换能器'的结构由与二价抑制剂为GST改性的抗PDGF的DNA适体。因此,该合成剂属于家族分子与结合位点不同的蛋白质,二聚的5-7如化学诱导剂(的CID)8-10并且还基于寡核苷酸的合成分子偶联物的组蛋白粘合剂。11-21
这种系统的设计基础的一般原理,本文描述和提供了用于合成和测试该常规酶测定“换能器”的功能的详细协议。这项工作是为了便于开发这一类,其可以用于介导的细胞内蛋白质 – 蛋白质的通信,因此,以诱导人工细胞信号传导途径的额外的“换能器”。
图1示意性地描述了合成“化学传感器”,可以调解不自然的蛋白质-蛋白质通信的工作原理。在该图中,“化学传感器”,它集成了普罗特合成粘合剂EINS I和II(粘合剂I和II),使蛋白质二触发蛋白I的催化活性,这不是其天然结合伴侣。在不存在蛋白Ⅱ的,换能器结合的酶(蛋白质I)中的催化位点并抑制其活性( 图1,状态ⅱ)。该'换能器'的蛋白II的结合,然而,促进粘合剂I和蛋白II的表面( 图1,状态ⅲ),这降低了它的朝向蛋白I.亲和力结果,的有效浓度之间的相互作用的'在溶液中自由'换能器被缩小,这导致所述换能器蛋白的解离我复杂和蛋白I的活化( 图1,状态ⅳ)。两者合计,这些步骤突出的高效'换能器'的设计基础三个基本原则:(1)一个“换能器”应具有特定的粘合剂对于每个蛋白质靶的,(2)的相互作用betwe烯粘合剂II和蛋白II应该比粘合剂I和蛋白I,和(3)的粘合剂之间的相互作用我必须能够与蛋白Ⅱ的表面相互作用更强。这最后的原则并不一定要求粘合剂我单独将具有朝向蛋白质二高亲和力和选择性。相反,它是根据我们最近的研究这表明使合成分子接近的蛋白质是可能促进该分子与所述蛋白质的表面之间的相互作用。19,22,23
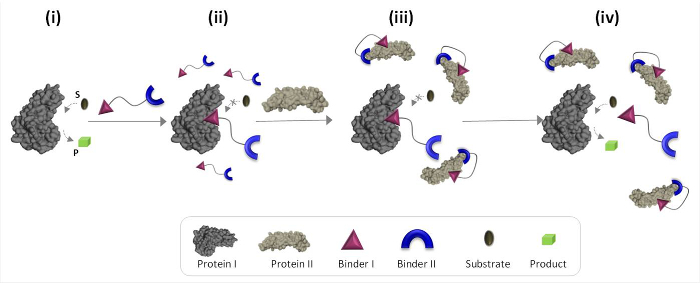
图1:的“化学换能器”的操作原则当“化学传感器”被添加到活性蛋白I(状态i),它通过粘结剂结合至其活性位点I和抑制其活性(状态II)。在蛋白Ⅱ的存在,然而,未结合的'化学吨ransducer'通过粘合剂二,这促进了粘合剂I和蛋白II的表面之间的相互作用与蛋白质II相互作用。这引起粘结剂I-II蛋白质相互作用降低了有效浓度粘结剂我的,这导致了“transducer'蛋白解离我复杂的蛋白质我复活(国IV)。 请点击此处查看该图的放大版本。
Protocol
Representative Results
Discussion
We presented a method for designing and testing of a ‘chemical transducer’ that can induce artificial communication between two naturally unrelated proteins, GST and PDGF, without modifying the native proteins. The unnatural GST-PDGF communications could be detected in real time by using enzymatic assays that follow the changes in the activity of GST in the presence of the ‘chemical transducer’ and increasing the concentrations of PDGF. In addition to detecting the activation of GST by PDGF, these assays were used to fol…
Disclosures
The authors have nothing to disclose.
Acknowledgements
这项研究是由密涅瓦基金会,HFSP组织和欧洲研究理事会资助项目(开始格兰特338265)。
Materials
| 1-chloro-2,4-dinitrobenzene | Sigma-Aldrich | 237329 | |
| Acetic acid | Bio Lab | 01070521 | |
| Acetnitrile | J.T.Baker | 9017-03 | |
| Ascorbic acid | Sigma-Aldrich | A4544 | |
| Copper(II) Sulfate pentahydrate | Merck-Millipore | 102790 | |
| Dimethyl sulfoxide | Merck-Millipore | 802912 | |
| Dulbecco's Phosphate Buffered Saline | Biological Industries | 02-023-5A | |
| Ethacrynic acid | Tokyo Chemical Industry Co. Ltd | E0526 | |
| Glutathione-s-transferase M1-1 | Israel Structural Proteomics Center (Weizmann Institute of Science, Rehovot, Israel) | ||
| JS-K | Sigma-Aldrich | J4137 | |
| L-glutathione reduced | Sigma-Aldrich | G4251 | |
| Magnesium Chloride | J.T.Baker | 0162 | |
| nitrate/nitrite colorimetric assay kit | Cayman Chemical | 780001 | |
| Oligonucleotides | W. M. Keck Foundation Biotechnology at Yale University | custom order | |
| PDGF-BB | Israel Structural Proteomics Center (Weizmann Institute of Science, Rehovot, Israel) | ||
| TBTA | Sigma-Aldrich | 678937 | |
| Triethylamine | Sigma-Aldrich | T0886 | |
| Desalting column | GE Healthcare | illustra MicroSpin G-25 Columns | |
| HPLC | Waters | 2695 separation module | |
| HPLC column | Waters | XBridgeTM OST C18 column (2.5 μM, 4.6 mm × 50 mm) | |
| HPLC column | Waters | XBridgeTM OST C18 column (2.5μM, 10 mm × 50 mm) | |
| Plate reader | BioTek | synergy H4 hybrid |
References
- Hunter, T. Signaling—2000 and Beyond. Cell. 100, 113-127 (2000).
- Levitzki, A., Klein, S. Signal transduction therapy of cancer. Mol Aspects Med. 31, 287-329 (2010).
- Peri-Naor, R., Motiei, L., Margulies, D. Artificial signal transduction therapy: a futuristic approach to disease treatment. Future Med. Chem. 7, 2091-2093 (2015).
- Peri-Naor, R., Ilani, T., Motiei, L., Margulies, D. Protein-Protein Communication and Enzyme Activation Mediated by a Synthetic Chemical Transducer. J. Am. Chem. Soc. 137, 9507-9510 (2015).
- Corson, T. W., Aberle, N., Crews, C. M. Design and Applications of Bifunctional Small Molecules: Why Two Heads Are Better Than One. ACS Chem. Biol. 3, 677-692 (2008).
- Rutkowska, A., Schultz, C. Protein Tango: The Toolbox to Capture Interacting Partners. Angew. Chem. Int. Ed. 51, 8166-8176 (2012).
- Meyer, C., Köhn, M. A Molecular Tête-à-Tête Arranged by a Designed Adaptor Protein. Angew. Chem. Int. Ed. 51, 8160-8162 (2012).
- Klemm, J. D., Schreiber, S. L., Crabtree, G. R. Dimerization as a Regulatory Mechanism in Signal Transduction. Annu. Rev. Immunol. 16, 569-592 (1998).
- DeRose, R., Miyamoto, T., Inoue, T. Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 465, 409-417 (2013).
- Gestwicki, J. E., Marinec, P. S. Chemical control over protein-protein interactions: beyond inhibitors. Comb. Chem. High Throughput. Screen. 10, 667-675 (2007).
- Battle, C., Chu, X., Jayawickramarajah, J. Oligonucleotide-based systems for input-controlled and non-covalently regulated protein binding. Supramol. Chem. 25, 848-862 (2013).
- Diezmann, F., Seitz, O. DNA-guided display of proteins and protein ligands for the interrogation of biology. Chem. Soc. Rev. 40, 5789-5801 (2011).
- Röglin, L., Ahmadian, M. R., Seitz, O. DNA-Controlled Reversible Switching of Peptide Conformation and Bioactivity. Angew. Chem. Int. Ed. 46, 2704-2707 (2007).
- Röglin, L., Altenbrunn, F., Seitz, O. DNA and RNA-Controlled Switching of Protein Kinase Activity. ChemBioChem. 10, 758-765 (2009).
- Harris, D. C., Chu, X., Jayawickramarajah, J. DNA-Small Molecule Chimera with Responsive Protein-Binding Ability. J. Am. Chem. Soc. 130, 14950-14951 (2008).
- Harris, D. C., Saks, B. R., Jayawickramarajah, J. Protein-Binding Molecular Switches via Host-Guest Stabilized DNA Hairpins. J. Am. Chem. Soc. 133, 7676-7679 (2011).
- Kim, Y., Cao, Z., Tan, W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc. Nat. Acad. Sci. U.S.A. 105, 5664-5669 (2008).
- Han, D., et al. A Logical Molecular Circuit for Programmable and Autonomous Regulation of Protein Activity Using DNA Aptamer-Protein Interactions. J. Am. Chem. Soc. 134, 20797-20804 (2012).
- Motiei, L., Pode, Z., Koganitsky, A., Margulies, D. Targeted Protein Surface Sensors as a Tool for Analyzing Small Populations of Proteins in Biological Mixtures. Angew. Chem. Int. Ed. 53, 9289-9293 (2014).
- Ranallo, S., Rossetti, M., Plaxco, K. W., Vallée-Bélisle, A., Ricci, F. A Modular, DNA-Based Beacon for Single-Step Fluorescence Detection of Antibodies and Other Proteins. Angew. Chem. Int. Ed. 54, 13214-13218 (2015).
- Franzini, R. M., et al. Identification of Structure-Activity Relationships from Screening a Structurally Compact DNA-Encoded Chemical Library. Angew. Chem. Int. Ed. 54, 3927-3931 (2015).
- Unger-Angel, L., et al. Protein recognition by bivalent, ‘turn-on’ fluorescent molecular probes. Chem. Sci. 6, 5419-5425 (2015).
- Nissinkorn, Y., et al. Sensing Protein Surfaces with Targeted Fluorescent Receptors. Chem. Eur. J. 21, 15981-15987 (2015).
- Huber, C. G., Oefner, P. J., Bonn, G. K. High-Resolution Liquid Chromatography of Oligonucleotides on Nonporous Alkylated Styrene-Divinylbenzene Copolymers. Anal. Biochem. 212, 351-358 (1993).
- Lyon, R. P., Hill, J. J., Atkins, W. M. Novel class of bivalent glutathione S-transferase inhibitors. Biochemistry. 42, 10418-10428 (2003).
- Battle, C., Chu, X., Jayawickramarajah, J. Oligonucleotide-based systems for input-controlled and non-covalently regulated protein binding. Supramol. Chem. , 1-16 (2013).

