鸡颅神经嵴细胞培养物的制备及形态学分析
Summary
这种多功能方案描述了通过从鸡胚胎中切除颅神经褶皱来分离前迁移神经嵴细胞(NCC)。在铺板和孵育时,迁移性NCC从神经折叠外植体中出现,允许在简化的2D环境中评估细胞形态和迁移。
Abstract
在脊椎动物发育过程中,神经嵴细胞(NCC)广泛迁移并分化成各种细胞类型,这些细胞类型有助于颅面骨骼和周围神经系统等结构。虽然在3D胚胎的背景下了解NCC迁移至关重要,但在2D培养中分离迁移细胞有助于可视化和功能表征,补充胚胎研究。本协议展示了一种分离鸡颅神经褶皱以产生原代NCC培养物的方法。迁移性NCC来自接种在纤连蛋白包被底物上的神经折叠外植体。这导致分散的、粘附的NCC群体,可以通过染色和定量形态学分析进行评估。这种简化的培养方法适应性强,可以与其他技术结合使用。例如,NCC迁移和迁移行为可以通过延时成像进行评估,或者通过包括抑制剂或基因表达的实验操作(例如,DNA,吗啉诺或CRISPR电穿孔)来询问功能。由于其多功能性,该方法为研究颅骨NCC的发展提供了一个强大的系统。
Introduction
神经嵴细胞(NCC)是脊椎动物胚胎中的瞬时细胞群。NCC 指定在神经板的边界,并经历上皮到间充质的过渡 (EMT) 以从背神经管迁移1。EMT后,NCC广泛分散在整个胚胎中,最终分化并促成各种结构,包括颅面骨骼,心脏流出道和大部分周围神经系统2。细胞极性、细胞骨架和粘附特性的变化是这种从迁移性细胞群向迁移性细胞群转变的基础3.研究NCC EMT和迁移提供了对细胞运动基本机制的见解,并为预防和治疗出生缺陷和癌症转移的努力提供了信息。
虽然体内分析对于理解胚胎环境中的NCC发育过程至关重要,但体外方法提供了视觉和物理可及性,促进了其他实验途径。在简化的 2D 环境中,可以评估 NCC 形态、细胞骨架结构和迁移距离。此外,遗传或可溶性因子扰动对运动NCC迁移行为的影响可以分析4,5,6,7,8,9,10。此外,可以收集、汇集分离的迁徙或迁移性 NCC 并用于高通量方法,以通过蛋白质组、转录组和表观基因组分析研究 NCC 的发育调控7,11。虽然有方法可用于从各种发育模式生物制备颅骨NCC12,13,14,但本文展示了那些首次学习从鸡胚胎培养颅骨NCC的方法的机制。
目前的协议描述了一种用于制备鸡颅NCC培养物的通用技术(图1)。由于NCC很容易从外植的神经褶皱迁移到培养基质上,因此雏鸡NCC自然地与胚胎组织分离,并且很容易产生原代培养物。随着中脑NCC从颅神经褶皱中大量迁移(与躯干15中长时间的逐细胞分层相反),这些培养物主要由迁移性颅神经嵴细胞组成,初始神经褶皱切除为迁移性NCC提供了一种收集方法。详细介绍了解剖和培养鸡颅神经褶皱的基本方法,并提供了该方法的不同应用和变化的建议。
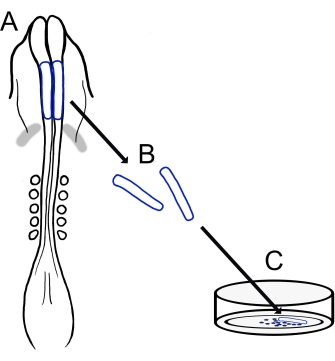
图 1:小鸡颅神经褶皱培养方案的示意图。 (A,B)从具有五个体细胞的雏鸡胚胎中切除颅神经褶皱(以蓝色轮廓显示)(如A的背视图所示)。灰色带,心脏新月形。(C)当接种在纤连蛋白上时,迁移性神经嵴细胞从神经褶皱中出现并分散到基质上。 请点击此处查看此图的大图。
Protocol
Representative Results
Discussion
这里描述的技术提供了一种适应性强的方法来分离雏鸡神经褶皱并对其进行铺板以创建迁徙性颅 NCC 培养物。这些培养物为轻松分析雏鸡NCC迁移和形态提供了简化的2D条件,可以补充卵巢成像方法中更具技术挑战性的24,25,26。虽然这种体外方法相对简单,但一致的结果取决于高质量的卵子和试剂。此外,由于培?…
Disclosures
The authors have nothing to disclose.
Acknowledgements
我们感谢Corinne A. Fairchild和Katie L. Vermillion,他们参与了我们版本的小鸡颅神经褶皱培养方案的开发。
Materials
| AxioObserver equipped with an LSM710 confocal scan head controlled by ZEN 3.0 SR software | Zeiss | Used alpha Plan-Apochromat 100x/1.46 Oil DIC M27 objective | |
| CaCl2 | Sigma-Aldrich | C3306 | |
| Chamber dishes (glass bottom, single or divided) | MatTek; Cell Vis | P35G-1.5-14-C (MatTek) X000NOJQGX (Cellvis) X000NOK1OJ (Cellvis) |
Single chamber 35 mm or 4 chamber 35 mm |
| Cover glass | Carolina Biological Supply Company | 633029, 633031, 633033, 633035, 633037 | circles, 0.13–0.17 mm thickness, available in 12-25 mm diameter |
| DMEM/F12 | ThermoFisher Scientific | 11320033 | Alternative for L15 media |
| Egg incubator | Sportsman | 1502 | |
| FBS | Life Technologies | 10437-028 | |
| Fibronectin | Fisher Scientific | CB-40008A | |
| Filter paper | Whatman | grade 3MM chromatography | |
| Forceps (blunt) | Fisher Scientific; Thomas Scientific | 08-890 (Fisher);1141W97 (Thomas) | |
| Forceps (fine) | Fine Science Tools | 11252-20 | Dumont #5 |
| Image J | https://fiji.sc/ | Free image analysis software | |
| KCl | Sigma-Aldrich | P3911 | |
| KH2PO4 | Sigma-Aldrich | P0662 | |
| L15 media | Invitrogen | 11415064 | |
| L-glutamine | Invitrogen | 25030 | |
| Mounting Media (Vectashield or ProLong Gold) | Vector Laboratories; Thermofisher Scientific | H-1700 (Vectashield); P36930 (ProLong Gold) | |
| Na2HPO4 | Sigma-Aldrich | S9638 | |
| NaCl | Sigma-Aldrich | S9888 | |
| Paraformaldehyde | Sigma-Aldrich | P6148 | |
| Penicillin/streptomycin | Life Technologies | 15140-148 | 10,000 Units/mL Penicillin; 10,000 mg/mL Streptomycin |
| Petri Dishes | VWR (or similar) | 60 mm, 100 mm | |
| Phalloidin | Sigma-Aldrich | P1951 | multiple flurophores available |
| Pin holder | Fine Science Tools | 26016-12 | For tungsten needle (alternative for spring scissors) |
| Scissors (dissection) | Fine Science Tools | 14061-10 | |
| Spring Scissors | Fine Science Tools | 15000-08 | 2.5 mm cutting edge (alternative for tungsten needle) |
| Sylgard | Krayden | Sylgard 184 | |
| Syringe Filters | Sigma-Aldrich | SLGVM33RS | Millex-GV Syringe Filter Unit, 0.22 µm, PVDF, 33 mm, gamma sterilized |
| Tissue culture dishes | Sarstedt | 83-3900 | 35 mm culture dishes for bulk neural fold cultures |
| Triton X-100 | Sigma-Aldrich | X100 | |
| Tungsten wire | Variety of sources | 0.01" diameter for tungsten needle (alternative for spring scissors) |
References
- Pla, P., Monsoro-Burq, A. H. The neural border: Induction, specification and maturation of the territory that generates neural crest cells. Developmental Biology. 444, 36-46 (2018).
- Tang, W., Bronner, M. E. Neural crest lineage analysis: From past to future trajectory. Development. 147 (20), (2021).
- Piacentino, M. L., Li, Y., Bronner, M. E. Epithelial-to-mesenchymal transition and different migration strategies as viewed from the neural crest. Current Opinion in Cell Biology. 66, 43-50 (2020).
- McLennan, R., et al. Neural crest cells bulldoze through the microenvironment using Aquaporin 1 to stabilize filopodia. Development. 147 (1), 185231 (2020).
- Carmona-Fontaine, C., et al. Complement fragment C3a controls mutual cell attraction during collective cell migration. Developmental Cell. 21 (6), 1026-1037 (2011).
- Giovannone, D., et al. Slits affect the timely migration of neural crest cells via robo receptor. Developmental Dynamics. 241 (8), 1274-1288 (2012).
- Vermillion, K. L., Lidberg, K. A., Gammill, L. S. Cytoplasmic protein methylation is essential for neural crest migration. Journal of Cell Biology. 204 (1), 95-109 (2014).
- Yang, X., Li, J., Zeng, W., Li, C., Mao, B. Elongator Protein 3 (Elp3) stabilizes Snail1 and regulates neural crest migration in Xenopus. Scientific Reports. 6 (1), 1-9 (2016).
- Gonzalez Malagon, S. G., et al. Glycogen synthase kinase 3 controls migration of the neural crest lineage in mouse and Xenopus. Nature Communications. 9 (1), 1-15 (2018).
- Bhattacharya, D., Azambuja, A. P., Simoes-Costa, M. Metabolic reprogramming promotes neural crest migration via yap/tead signaling. Developmental Cell. 53 (2), 199-211 (2020).
- Jacques-Fricke, B. T., et al. Profiling NSD3-dependent neural crest gene expression reveals known and novel candidate regulatory factors. Developmental Biology. 475, 118-130 (2021).
- Bronner-Fraser, M., García-Castro, M. Chapter 4 manipulations of neural crest cells or their migratory pathways. Methods in Cell Biology. 87, 75-96 (2008).
- Milet, C., Monsoro-Burq, A. H. Dissection of xenopus laevis neural crest for in vitro explant culture or in vivo transplantation. Journal of Visualized Experiments. (85), e51118 (2014).
- Malagon, S. G. G., et al. Dissection, culture and analysis of primary cranial neural crest cells from mouse for the study of neural crest cell delamination and migration. Journal of Visualized Experiments. (152), e60051 (2019).
- Theveneau, E., Mayor, R. Neural crest delamination and migration: From epithelium-to-mesenchyme transition to collective cell migration. Developmental Biology. 366 (1), 34-54 (2012).
- Conrad, G. W., Bee, J. A., Roche, S. M., Teillet, M. A. Fabrication of microscalpels by electrolysis of tungsten wire in a meniscus. Journal of Neuroscience Methods. 50 (1), 123-127 (1993).
- Hamburger, V., Hamilton, H. L. A series of normal stages in the development of the chick embryo. Journal of Morphology. 88 (1), 49-92 (1951).
- Gammill, L. S., Jacques-Fricke, B., Roffers-Agarwal, J. Embryological and genetic manipulation of chick development. Methods in Molecular Biology. 1920, 75-97 (2019).
- Vandekerckhove, J., Deboben, A., Nassal, M., Wieland, T. The phalloidin binding site of F-actin. The EMBO Journal. 4 (11), 2815-2818 (1985).
- Bronner-Fraser, M. Analysis of the early stages of trunk neural crest migration in avian embryos using monoclonal antibody HNK-1. Developmental Biology. 115 (1), 44-55 (1986).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Soille, P., Vincent, L. Determining watersheds in digital pictures via flooding simulations. Visual Communications and Image Processing ’90: Fifth in a Series. (1360), 240-250 (1990).
- Haupt, A., Minc, N. How cells sense their own shape – mechanisms to probe cell geometry and their implications in cellular organization and function. Journal of Cell Science. 131 (6), (2018).
- Ezin, M., Fraser, S. Chapter 11 time-lapse imaging of the early avian embryo. Methods in Cell Biology. 87, 211-236 (2008).
- Kulesa, P. M., Bailey, C. M., Cooper, C., Fraser, S. E. In ovo live imaging of avian embryos. Cold Spring Harbor Protocols. 5 (6), (2010).
- McKinney, M. C., Kulesa, P. M. Live imaging of the neural crest cell epithelial-to-mesenchymal transition in the chick embryo. Methods in Molecular Biology. 2179, 107-114 (2021).
- Gustafson, C. M., Roffers-Agarwal, J., Gammill, L. S. Chick cranial neural crest cells release extracellular vesicles that are critical for their migration. Journal of Cell Science. , (2022).
- Williams, R., Sauka-Spengler, T. Ex ovo electroporation of early chicken embryos. STAR Protocols. 2 (2), 100424 (2021).
- Moulton, J. D. Using morpholinos to control gene expression. Current Protocols in Nucleic Acid Chemistry. 68 (1), 4-30 (2017).
- Gandhi, S., et al. A single-plasmid approach for genome editing coupled with long-term lineage analysis in chick embryos. Development. 148 (7), (2021).
- Williams, R. M., et al. Reconstruction of the Global Neural Crest Gene Regulatory Network In Vivo. Developmental Cell. 51 (2), 255-267 (2019).

