면역침전반응 기반 기술: 아가로스 비즈를 사용한 내인성 단백질의 정제
English
Share
Overview
출처: 수잔나 C. 시슬러1,토냐 J. 웹1
1 미생물학 및 면역학학과, 메릴랜드 대학교 볼티모어, MD 21201
면역 침전물(IP, ‘풀다운’ 분석이라고도 함)은 다양한 분야에서 응용 분야를 가지고 있는 널리 사용되는 기술이다. 1984년에 처음 구상된 이 요리는 1988년(1, 2)에 정제되었다. IP의 근본적인 목표는 그 단백질에 대하여 항체를 사용하여 특정 단백질의 정제 그리고 격리입니다. 단어 “면역”은 “강수량”이라는 단어가 용액에서 특정 물질을 당기는 것을 의미하는 동안 항체의 사용을 지칭한다. 표적 단백질은 내인성 또는 재조합일 지도 모릅니다. 대부분의 재조합 단백질에는 후속 정화를 단순화하기 위해 에피토프 태그(즉, myc 또는 flag)가 부착되어 있습니다. 전형적으로, 재조합 에피토프 태그에 대한 항체가 매우 강하고 효과적이기 때문에 재조합 단백질 IP를 최적화하는 것이 더 쉽습니다. 내인 성 단백질에 대한 항체는 매우 가변적 인 효능을 가지고 있어 이러한 IP를 최적화하기가 훨씬 더 어려워지게됩니다. 면역 침전 후 필요한 단계는 정화의 검증이다. 격리된 단백질은 SDS-PAGE를 사용하여 해결되고 그 후 서양 얼룩에 의한 순도를 위해 조사됩니다(그림 1). 중요한 대조군은 정확한 단백질의 당김을 확인하기 위해 서양 블롯 동안 다른 항체를 사용하는 것이다. IP와 후속 기술의 조합은 강력한 분석 도구입니다. 정제 후의 목표는 NMR, 질량 분광법 및 시험관 내 분석, 또는 단백질의 상호 작용 파트너(즉, 단백질, DNA, RNA)의 분석(즉, 단백질, DNA, RNA) (3, 4, 5)에 의해 단백질 자체의 특성화일 수 있다.
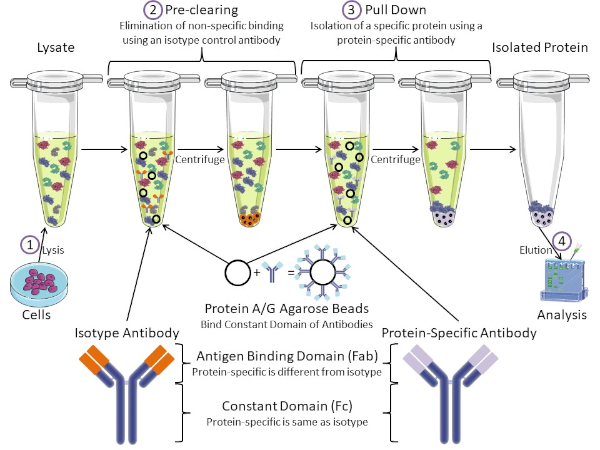
그림 1: 면역 침전 절차의 개요. 면역 침전은 항체를 사용하여 특정 단백질의 격리이다. 세포로부터 용재를 생산한 후, 두 가지 주요 단계-사전 정리 및 풀다운이 있습니다. 사전 클리어링 단계 동안, 세포-lysates는 isotype 대조군 항체를 사용하여 비특이적으로 항체에 결합하는 단백질의 미리 지워집니다. 당기면, 표적 단백질은 단백질 특이적 항체를 사용하여 아래로 당겨졌다. 고립 된 단백질은 서양 얼룩에 의해 분석됩니다. 동형 항체 및 단백질 특이적 항체는 동일한 상수 도메인을 가지지만, 상이한 항원 결합 도메인이 있다. 이 프로토콜의 핵심 성분은 항체의 일정한 도메인을 결합하는 단백질 A/G 아가로즈 구슬입니다 – 표적 단백질의 면역 침전을 허용합니다. 이 그림의 더 큰 버전을 보려면 여기를 클릭하십시오.
항체는 다른 형태의 단백질 정제(즉, 니켈 친화 컬럼 정제)와 구별되는 면역 침전의 핵심 성분이다. 항체는 특정 단백질 에피토프를 인식할 수 있는 B 세포에 의해 만들어진 분자입니다. 항체에는 상수(Fc) 및 항원 결합(Fab)(도 1)의 두 가지 영역이 있습니다. 상수 도메인은 항체의 유형을 식별하고 생체 내에서기능을 지시한다. 일반적으로 IP에 사용되는 항체의 상수 영역은 마우스, 쥐 또는 토끼 IgG입니다. 항체의 항원 결합 부는 특정 단백질의 특정 에피토프를 인식한다. 항체는 단백질이 변성될 때 존재하지 않을 수 있는 접힌 단백질에 대한 전형체를 인식할 수 있고 그 반대의 경우도 마찬가지입니다. 따라서, 에피토프의 가용성은 단백질 접이식에 달려 있습니다 – IP에 대한 항체 및 조건을 선택할 때 고려해야 할 중요한 요소를 식별합니다.
대핵계와 진핵계에는 항체 결합 단백질이 있습니다. 진핵 계통에서, 목적은 근생 계통에 있는 동안 박테리아로부터 면역 보호입니다, 목적은 면역 계통으로부터 보호입니다. 항체 결합 단백질은 두 가지 방법으로 IP 방법론에 영향을 미칩니다. 첫째, 항체를 결합하는 단백질의 용액을 제거하는 데 필요한 사전 정리 단계(도 1)가 있어 최종 제품에 비특이적 결합을 줄입니다. 이 단계는 단백질 특이적 항체와 는 다른 항체 결합 도메인과 동일한 상수 영역을 가지고 있는 동종형 항체를 사용합니다. 세균성 항체 결합 단백질은 이 방법의 두 번째 핵심 성분이다. 단백질 특이적 항체가 표적 단백질을 결합한 후, 항체: 단백질 복합체는 아래로 당겨져야 한다(도 1). 단백질 A, G 및 L은 항체의 일정한 도메인을 결합하는 세균성 단백질입니다. 박테리아는 면역 계통을 전복하기 위하여 이것을 사용하는 동안, 연구원은 쉬운 항체 정화를 위한 이 시스템을 공동 선택했습니다, 그리고 사전 정리 및 풀다운 단계 둘 다 도중 이용됩니다. 이 단백질은 다른 종 및 다른 일정한 도메인 하위형에 대한 상이한 결합 친화성을 가지고 있습니다 – IP를 위한 조건을 선택할 때 고려해야 할 또 다른 요인. 많은 회사에서 단백질 A/G 라벨아가로즈 구슬(그림 1), 미리 만들어진 스핀 컬럼 또는 수지를 판매하여 컬럼을 만듭니다. 일반적으로 구슬과 스핀 컬럼은 더 작은 샘플 크기에 사용되는 반면 수지는 대량 정화에 사용됩니다.
이 실험실 운동에서, 우리는 단백질 A/G 플러스 아가로즈 구슬 기지를 둔 기본적인 면역 침전 기술을 사용하여, 1 차적인 뮤린 흉구에서 내인성 단백질 c-myc를 정화하는 방법을 보여줍니다. 프로토콜은 세포 용해 준비에서 시작하고 서양 얼룩 분석을 사용하여 성공적인 단백질 풀다운의 검증으로 끝납니다.
Procedure
Results
The results of the procedure detailed above are shown in Figure 2. From left to right, the lanes contain the control group (isotype), the test group (c-myc), the pre-cleared lysate (lysate), and the molecular weight ladder (ladder). The 25 and 75 kDa ladder bands are marked. The two prominent bands at ~25 kDa and 50 kDa are the light and heavy chain of the binding antibody, respectively and are non-specific to the IP or the samples. c-myc protein which runs around 67kDa on Western blots and is usually visible just below the 75 kDa ladder band. In this blot, the c-myc band is visible in the second lane, but absent in the first lane, indicating that the IP antibody successfully pulled down c-myc. There is no visible band in the pre-cleared lysate lane, suggesting that this protein has low endogenous expression levels.
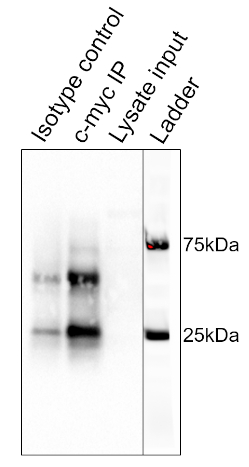
Figure 2: Results of a Western Blot Analysis, used to assess the purification of c-myc by immunoprecipitation. A band at 67 kDa, corresponding to c-myc, is visible in the anti-c-myc lane, but not the isotype control lane. Note that c-myc levels were not high enough to be visualized in the lysate lane. Please click here to view a larger version of this figure.
Applications and Summary
In short, immunoprecipitation is the isolation of a specific protein using an antibody. In this example, the results of the immunoprecipitation were analyzed by Western blot to assess the purity. The isolated protein could be used in a number of applications afterwards including: NMR for protein structure, Mass Spectrometry for amino acid sequence, or in vitro assays for enzymatic characterization. IPs can also characterize the interacting partners of proteins. For instance, following isolation, DNA or RNA could be isolated for sequencing. Co-immunoprecipitations assess protein-protein interactions. When the target protein is pulled down during an IP, interacting proteins can also be pulled down. These interacting partners can be assessed by mass spectrometry and Western blot. Immunoprecipitation is a powerful technique for studying protein biology.
References
- Olliver, C. L. and Boyd, C. D. (1984). Immunoprecipitation of In Vitro Translation Products with Protein A Bound to Sepharose. In J. M. Walker (eds), Nucleic Acids. Methods in Molecular Biology (pp. 157-160). New Jersey: Humana Press.
- Thurston, C. F. and Henley, L. F. (1988). Direct Immunoprecipitation of Protein. In J. M. Walker (eds), New Protein Techniques. Methods in Molecular Biology (pp. 149-158). New Jersey: Humana Press.
- Anderson, N. G. (1998). Co-immunoprecipitation: Identification of Interacting Proteins. In R. A. Clegg (eds), Protein Targeting Protocols.Methods in Molecular Biology (pp. 35-45). New Jersey: Humana Press.
- Jackson, D. I. and Dickson, C. (1999). Protein Techniques: Immunoprecipitation, In Vitro Kinase Assays, and Western Blotting. In P.T. Sharpe and I. Mason (eds), Molecular Embryology. Methods in Molecular Biology (pp. 699-708). New Jersey: Humana Press.
- Trieu, E. P. and Targoff, I. N. (2015). Immunoprecipitation: Western Blot for Proteins of Low Abundance. In B. Kurien and R. Scofield (eds), Western Blotting. Methods in Molecular Biology (pp. 327-342). New York, NY: Humana Press.
Transcript
Immunoprecipitation, or IP, is a widely used technique to isolate a protein of interest from a cell or tissue lysate or a body fluid for protein characterization or to investigate protein-protein interactions.
The process begins with an antibody, which has a high affinity and specificity for the target protein. This antibody is mixed with the sample, allowing antibody-target complexes to form. Any protein bound to the target protein also gets indirectly attached to the antibody in the process. Next, the solution is incubated with agarose beads, conjugated to a bacterial protein, which has a strong affinity for the constant region of antibodies. The bacterial protein binds to the antibody and connects the antibody- target complexes to the beads. Then, the solution is centrifuged to precipitate the beads, thereby extracting the entire complex containing the binding antibody, the target protein, and any interacting proteins. Finally, the bound proteins are extracted from the beads and released from each other and are used for further analysis by techniques such as Western blotting.
Several variations of different parts of this technique are commonly used, like pre-clearing, using peptide tags or magnetic beads, or analyzing other non-protein binding partners. IP can be preceeded by a pre-clearing step, to remove non-specific antibody-binding proteins in the sample and minimize background. This involves first incubating the sample with isotype control antibodies, allowing them to bind to these proteins, and then using agarose beads to precipitate the complexes. The sample is then ready to proceed to the actual IP.
Peptide tags are useful if a specific antibody is not available for IP. Here, the target protein can be genetically modified to contain a peptide epitope tag and an antibody against the tag is able to pull out the protein of interest. Magnetic beads are often used instead of agarose to precipitate the target. After binding to the antibody-target complex, the sample tube is placed in a strong magnetic field, which extracts the beads from the solution. This eliminates the need for centrifugation and improves speed and convenience.
Immunoprecipitation is also used for studying DNA or RNA binding proteins and are known as chromatin immunoprecipitation and RNA immunoprecipitation, respectively. These variations are useful for troubleshooting and adapting the method for different experimental applications. In this video, you will observe how to pre-clear a cell lysate and perform immunoprecipitation to extract a protein of interest, followed by Western blot analysis to validate the experiment.
To begin, place the pre-collected cells in a microcentrifuge and spin at 13 thousand rpm for three minutes. Following the spin, remove the supernatant and then resuspend the cells in 500 microliters of lysis buffer RIPA with PMSF. Now, disrupt the cells using a few quick pulses with a vortex and then aspirate the lysate a few times with a 25 gauge needle attached to a syringe, taking care to avoid creating bubbles. Place the cells on ice for 15 minutes. After incubating the samples on ice, centrifuge the lysate for 15 minutes at four degrees celsius.
Label a new 1.5 milliliter microcentrifuge tube. Following the spin, transfer the supernatant to the freshly labeled tube and discard the pellet. Next, pre-clear the lysate of contaminants that bind non-specifically to either the agarose beads or the primary antibody by adding 20 microliters of the Protein A/G PLUS-agarose beads and one microgram of an isotype control antibody to the lysate, which in this example is a mouse IgG1 isotype control. Incubate the tube on a rotator in a cold room for 30 minutes. After rotating the lysate in the cold room for 30 minutes, centrifuge the sample at 3200 rpm for 30 seconds at four degrees celsius. Remove the tube from the centrifuge and transfer the pre-cleared supernatant to a fresh labeled 1.5 milliliter microcentrifuge tube. Discard the pellet.
Now, determine the protein concentration of the cell lysate by performing a Bradford assay. Label seven 1. 5-milliliter microcentrifuge tubes one through six and sample and aliquot 1000 microliters of the Bradford reagent into each tube. Six of the tubes will be used to make a standard curve by adding various amounts of known quantities of BSA to each tube. The amounts to add are listed in this table. In the seventh sample tube, add one microliter of the pre-cleared lysate. Place 200 microliters from each of the seven tubes into individual wells of a flat-bottom 96-well plate, repeating each sample in triplicate so that there are three columns of seven samples. Read the plate on a plate reader, using a wavelength of 595 nanometers. After creating a standard curve in Excel, calculate the protein concentration of the pre-cleared lysate.
Next, label two 1.5-milliliter microcentrifuge tubes- one as control and the other as test, which in this example, will be the c-myc antibody. Place 500 micrograms of the pre-cleared lysate into each of these tubes and then bring the total volume for each tube up to 500 microliters using lysis buffer. Next, add two micrograms of the anti-c-myc antibody to the test group tube. For the control, add two micrograms of the mouse IgG1 isotype control antibody. Once the antibodies are added to the tubes, place the samples on a rotator in a cold room and incubate for two hours. Now, add the agarose beads. To do this, it is recommended to cut off the end of a pipette tip and then, using this modified tip, add 200 microliters of the Protein A/G PLUS-agarose beads to each tube. Incubate the tubes on a rotator in the cold room overnight.
Following the incubation, remove the tubes from the rotator and spin the lysates in the microcentrifuge to pull down the beads. After the spin is complete, remove the tubes from the centrifuge and aspirate the supernatant from each tube. Next, wash the beads using 500 microliters of 1X Dulbecco’s PBS. Place the tubes in a microcentrifuge and spin down for 30 seconds at four degrees celsius. Following this, remove the supernatant. Repeat the wash and centrifuge steps one more time for a total of two times. Remove the tubes from the microcentrifuge and aspirate the buffer from each tube. Using gel loading tips, remove any left over buffer from the beads, keeping the beads on ice to elute the bound protein.
In this example, the protein is eluted into SDS-PAGE running buffer by boiling for Western blot analysis. To do this, resuspend the beads in 20 microliters of SDS-PAGE loading dye containing beta-mercaptoethanol, or BME. Boil the samples at 95 degrees celsius for five minutes to dissociate the immunocomplexes from the beads. Then, centrifuge the beads at maximum speed for 10 seconds at room temperature. Remove the tubes from the microcentrifuge and hold them in a rack at room temperature. Using gel loading tips, carefully pipette the samples from the beads and load them into wells of a 4 to 15% gradient SDS-PAGE gel. In addition to the samples, load a lane with a protein ladder as well as a lane with the pre-cleared lysate to serve as a loading control. Once the gel is loaded, run the gel at 100 volts.
After the dye front has reached the bottom of the gel, which should take approximately one hour, stop the gel and make a Western blot sandwich, ensuring that the PVDF membrane is between the gel and the cathode. Place the Western blot sandwich in the transfer apparatus and transfer the proteins on the gel to the membrane for one hour at 100 volts. After the transfer is complete, place the membrane in five milliliters of block to prevent the antibodies from binding non-specifically to the membrane. Rock at a low setting for an hour at room temperature. When the timer sounds, remove the blocking buffer. Add five milliliters of the blocking buffer with the detection antibody to the membrane. Here, an anti-c-myc antibody, that is different than the one used for the pull down, is used.
Incubate the blot over night, at four degrees celsius on a rocker at a low setting. Following the incubation, remove the antibody and blocking buffer. Wash the blot, using five milliliters of TBST for five minutes at room temperature, on a rocker at a low setting. This wash step should be repeated two to five times for a total of three to six washes, using fresh TBST for each wash. Add five milliliters of one to 1000 secondary antibody and blocking buffer to the blot. In this case, the secondary antibody is HRP-tagged anti-rabbit light chain. Incubate the blot on a rocker at a low setting for one our at room temperature. Next, remove the buffer and wash the blot with five milliliters of TBST. Incubate this wash on a rocker at a low setting for five minutes at room temperature. Repeat this wash for a total of six to 12 washes, each with a fresh five milliliters of TBST. Remove the final wash by first pouring the liquid off of the blot. Then, using tweezers, dab the edge of the blot on a laboratory wipe to remove any excess liquid and then place the blot in a fresh container. Next, cover the blot with 1X Chemiluminescent Detection Reagent and incubate for one minute.
Working quickly, dab the edge of the blot on a laboratory wipe to remove any excess detection reagent and then place the blot on the imaging surface of the Imager tray. Image using the Chemiluminescent program to capture multiple time points from 10 to 30 seconds. After the blot is imaged, choose an image with optimal band visibility and then export that image. Prior to moving the blot, use the Imager to take a picture of the blot to capture the location of the ladder. Then, export that image also. Finally, using a slide preparation software, such as PowerPoint, align the bands and ladder images to form one image.
This image shows the Western blot result for immunoprecipitation of the protein c-myc from thymocyte cells. From left to right, the lanes represent the isotype control, the c-myc IP, and the pre-cleared lysate input. The lane on the extreme right is a merged image of the molecular weight ladder. The strong band, at around 25 kilodaltons is from the light chain and the one at 50 kilodaltons is from the heavy chain of the binding antibody and are non-specific to the IP or the samples. C-myc runs around 67 kilodaltons on Western blots and is usually visible just below the 75 kilodalton ladder band. In this blot, the c-myc band is visible in the second lane but absent in the first lane, indicating that the IP antibody successfully pulled down c-myc. There is no visible band in the pre-cleared lysate lane, suggesting that this protein has low endogenous expression levels.
