Reconstruct Human Retinoblastoma In Vitro
Summary
We describe a method for generating human retinoblastoma (RB) by introducing biallelic RB1 mutations in human embryonic stem cells (hESC). RB cell lines could also be successfully cultured using the isolated RB in a dish.
Abstract
Human RB is pediatric cancer, which is lethal if no treatment is administered. As RB originates from cone precursors, which is relatively rare in rodent models, meanwhile regarding the interspecies differences between humans and rodents, a disease model derived from humans is more beneficial for uncovering the mechanisms of human RB and seeking the targets of therapy. Herein, the protocol describes the generation of two gene-edited hESC lines with a biallelic RB1 point mutation (RB1Mut/Mut) and an RB1 knockout mutation (RB1-/-), respectively. During the process of retinal development, the formation of RB is observed. The RB cell lines are also established by segregating from the RB organoids. Altogether, by differentiating the gene-edited hESC lines into the retinal organoids using a 2D and 3D combined differentiation protocol, we have successfully reconstructed the human RB in a dish and identified its cone-precursor origin. It would provide a helpful disease model for observing the retinoblastoma genesis, proliferation, and growth as well as further developing novel therapeutic agents.
Introduction
Human retinoblastoma (RB) is a rare, fatal tumor derived from the retinal cone-precursors1,2,3, is the most common type of intraocular malignancy in childhood4. Homozygous inactivation of RB1 gene is the initiating genetic lesion in RB5. However, mice with RB1 mutations fail to form the retinal tumor2. Although the mouse tumors could be generated with the combination of Rb1 mutations and other genetic modifications, they still lack the features of human RB6. Thanks to the development of retinal organoid differentiation, the hESC-derived RB could be obtained, displaying the characters of human RB1.
Numerous protocols for retinal organoid differentiation have been established in the past decade, including 2D7, 3D8, and a combination of 2D and 3D9. The method used here to generate the human RB is the consolidation of adherent culture and floating culture9. By differentiating the RB1 mutated hESC into retinal organoids, the formation of RB is detected at around day 45, and then it proliferates rapidly at around day 60. On day 90, isolation of RBs, and generation of the RB cell line is possible; furthermore, RB surrounds almost all retinal organoids at day 120.
hESC-derived RB is an innovative model for exploring the origin, tumorigenesis, and treatments for RB. In this protocol, the generation of gene-editing hESC, the differentiation of RB, and characterization for RB are described in detail.
Protocol
This study is approved by the institutional Ethics Committee of Beijing Tongren Hospital, Capital Medical University. H9 hESCs are obtained from the WiCell Research Institute.
1. Generation of RB1 mutated hESC
- CRISPR/Cas9 targeting vector for the knockout (KO) of RB1.
- Design a pair of sgRNA. For the ablation of RB1, target the first exon of this gene. The forward primer sequence is CACCGCGGTGGCGGCCGTTTTTCGG, and the reverse primer sequence is AAACCCGAAAAACGGCCGCCACCGC.
NOTE: For the specific RB1 mutation, a repaired template is also required. In this protocol, RB1-KO cell line is used as an example. - Digest 1 µg of pX330-U6-Chimeric BB-CBh-hSpCas9-2A-Puro with BbsI restriction enzymes (see Table of Materials) for 30 min at 37 °C by following the manufacturer's instructions.
- Purify the digested plasmids using a purification kit according to the manufacturers' instructions.
NOTE: Enzymatic reactions could be directly cleaned up without agarose gel using the purification kit (see Table of Materials). - Phosphorylate and anneal each pair of oligos using the T4 polynucleotide kinase (PNK) (Table of Materials) with the reaction comprising 1 µL of oligo1 (100 µM), 1 µL of oligo2 (100 µM), 1 µL of 10x T4 Ligation Buffer, 6.5 µL of ddH2O, and 0.5 µL of T4 PNK.
NOTE: The forward and reverse primers in step 1.1.1 are a pair of oligos. Phosphorylate and anneal the oligos in a thermocycler using the following parameters: 37 °C for 30 min; 95 °C for 5 min; ramp down to 25 °C at 5 °C per minute. - Dilute the phosphorylated and annealed oligos 200 times by adding 1 µL of oligo reagent to 199 µL of nuclease-free water at room temperature (RT).
- Set up the ligation reaction and incubate at RT for 10 min by mixing the following: 50 ng of Bbs1 digested plasmid from step 1.1.3, 1 µL of oligo duplex from step 1.1.5, 5 µL of 2x ligation buffer, 1 µL of ligase, and top up to 10 µL using nuclease-free water.
- Transform the ligation mixture and sequence the positive colonies for the next step.
NOTE: Use U6 forward or reverse primer for sequencing.- Thaw the competent cells on ice.
- Take 50 µL of the competent cells in a 1.5 mL microcentrifuge tube.
- Add 1 µL of the ligation mixture from step 1.1.6 into the competent cells.
- Mix gently by pipetting up and down the tube three times.
NOTE: Do not vortex. - Place the mixture on ice for 30 min.
- Heat shock the mixture at 42 °C for 90 s.
- Add 950 µL of room temperature Luria-Bertani (LB) broth without antibiotic to the tube.
- Place the tube in a shaker at 300 rpm at 37 °C for 60 min.
- Warm the LB plates (peptone, peptone from casein, sodium chloride, agar-agar, and ampicillin) to 37 °C in advance.
- Spread 50-100 µL of the cells and ligation mixture onto the plates at RT.
- Incubate the plates overnight at 37 °C.
- Pick up 12 colonies from the plate and transfer them to the LB medium with ampicillin. Place the medium on a shaker at 300 rpm for 60 min.
- Use 1 µL of the bacteria solution (from step 1.1.7.12) as template for PCR and select the positive colonies.
- Sequence the positive colonies by the U6 primers, use the colony with the designed sgRNA sequences in the next step.
- Extract the plasmid using a midi kit (see Table of Materials).
NOTE: The quality and the quantity of the plasmid using a mini kit are not enough for the following nucleofection experiment. Midi or maxi kit is more suitable than the mini kit.
- Design a pair of sgRNA. For the ablation of RB1, target the first exon of this gene. The forward primer sequence is CACCGCGGTGGCGGCCGTTTTTCGG, and the reverse primer sequence is AAACCCGAAAAACGGCCGCCACCGC.
- HESC culture and nucleofection.
NOTE: Prewarm all the reagents to RT.- Thaw 1 x 106 H9 cells into a pre-coated 6-well plate with 10 µM Y-27632 (ROCK inhibitor) and 2 mL of fresh ncEpic-hiPSC/hESC culture medium.
NOTE: Coat the 6-well plate with 1% Growth factor reduced basement membrane matrix for at least 30 min at 37 °C before use. - The next day, remove the supernatant, rinse the cells with 1x Dulbecco's Phosphate-Buffered Saline (DPBS), and then add 2 mL of fresh ncEpic-hiPSC/hESC culture medium.
- Change the medium every day with 2 mL of fresh ncEpic-hiPSC/hESC culture medium in the following 3-5 days until the cells grow to around 80% confluency.
- Passage once or twice to adjust the state of the hESC.
NOTE: The easiest way to identify the state of the hESC is the morphology. - Culture the undifferentiated H9 cells in a 6-well plate until they reach 80% confluency.
- Change the medium 2 h before the nucleofection with 2 mL of fresh ncEpic-hiPSC/hESC culture medium mixed with 10 µM of Y-27632 and precoat one well of a 12-well plate using 1% Growth factor reduced basement membrane matrix.
- Aspirate the medium and rinse with 1 mL of prewarmed DPBS.
- Remove the DPBS and incubate with 1 mL of cell dissociation enzyme (see Table of Materials) for 3.5 min at 37 °C.
- Add 1 mL of fresh ncEpic-hiPSC/hESC culture medium to the cells and pipette twice to make the H9 cells into a single cell suspension.
- Take out 100 µL of the mixture for cell counting and transfer the remaining to a 15 mL centrifuge tube containing another 2 mL of ncEpic-hiPSC/hESC culture medium inside.
- Centrifuge at 200 x g for 5 min. Remove the supernatant and resuspend around 2 x 106 cells into a 100 µL reaction volume. According to the manufacturer's protocol, optimize the volume for the cells, plasmids, nucleofector, and reaction conditions. See Table of Materials for the nucleofection system used in this protocol.
NOTE: Generally, bubbles are not allowed during the nucleofection. - After transfection, transfer the cells to the precoated 12-well plate, supplement with 1.5 mL of ncEpic-hiPSC/hESC culture medium and 10 µM of Y-27632, and then culture the cells in a 37 °C, 5% CO2 incubator.
- Change the medium daily. After 48 h, add 2 µg/mL of puromycin for cell selection around one week.
NOTE: Numerous cells die during the first 3 days. The remaining cells may proliferate and generate into colonies in the following week after the puromycin selection. In the selection week, ensure that the medium always contains 2 µg/mL of puromycin. - Pick the colonies manually under a microscope.
NOTE: The colony morphology is the same as the hES colony. Cut the colonies into pieces (2-6 pieces per colony) with a 10 µL white/normal pipette tip in the medium and transfer one colony into one precoated well of a 12-well plate. - First, Identify the RB1 mutations and the off-target situations (Table 1), and then pluripotency to characterize the RB1 knockout H9 cell line.
NOTE: Detect the RB1 mutations and the off-target situations using PCR and sanger sequencing. Identify the pluripotency markers by RT-PCR and Immunofluorescence.
- Thaw 1 x 106 H9 cells into a pre-coated 6-well plate with 10 µM Y-27632 (ROCK inhibitor) and 2 mL of fresh ncEpic-hiPSC/hESC culture medium.
2. Generation of human retinoblastoma
- Maintenance of RB1-KO hESC.
- Culture the gene-edited H9 cells in a Growth factor reduced basement membrane matrix coated 6-well plate with 2 mL of ncEpic-hiPSC/hESC culture medium and change the medium every day.
- Passage using EDTA buffer for 3.5 min at 37 °C or 5 min at RT.
NOTE: EDTA buffer is the mixture of 1x DPBS, 5 mM EDTA, and 0.9 mg/mL of NaCl.
- Retinal cell differentiation.
NOTE: Prepare Medium I before the differentiation. To prepare Medium I, mix 24.5 mL of Dulbecco's Modified Eagle Medium (DMEM)/Nutrient Mixture F-12(F12)-Glutamine (1x), 24.5 mL of Neuronal basal medium, 250 µL of 100x supplement A, 500 µL of 50x supplement B, 0.1 mM ß-mercaptoethanol, and 250 µL of 100x L-glutamine. Prewarm all the mixtures to RT before use.- Grow the RB1-KO hESC to 80% confluence in one well of a 6-well plate.
- Remove the medium and rinse the cells with 1 mL of 1x DPBS.
- Elevate the cell colonies using 1 mL of dispase buffer (see Table of Materials) for 5 min at 37 °C.
NOTE: Check the cell colonies under a microscope. Usually, it takes 5 min for the edge of the colonies to roll out. - Aspirate the dispase buffer and gently rinse the cells once with 1 mL of 1x DPBS.
- Add 1 mL of Medium I into the well.
- Cut the colonies into pieces (6-9 pieces per colony) with a 10 µL white/normal pipette tip in the medium.
NOTE: Generally, around 60%-70% of cells detach from the well. Use a cell scraper to scrape the remaining cells in the medium. - Harvest all the cells by centrifuging in a 15 mL tube at 200 x g for 5 min.
- Leave around 50 µL of the supernatant to disperse the cells in the medium, and then mix the cells with 250 µL of Growth factor reduced basement membrane matrix by gentle agitation.
NOTE: Keep the Growth factor reduced basement membrane matrix either at 4 °C or on the ice before use. If it is aliquoted and stored at -20 °C, place it at 4 °C overnight or at least 1 h before use. - Keep the 15 mL tube with the suspended cells into a 37 °C incubator for 20 min.
NOTE: Ensure that the cells and Growth factor reduced basement membrane matrix form a solidified gel. - Add 1 mL of Medium I into the 15 mL tube, and then lightly pipet the solidified gel two to three times to disperse the clump with a 1 mL pipette.
- Add 9 mL of Medium I and transfer the cell suspension into a 10 cm cell culture dish.
- Move the dish to a 37 °C incubator with 5% CO2 and set the day as day 0.
- Examine the cells using a microscope on day 1.
NOTE: Thousands of hollow cysts could be observed in the dish. On average, 3 to 4 cysts are observed in one piece of gel. - Change the medium on day 5. Collect the supernatant into a 15 mL tube, wait for the cysts to settle to the bottom, and then remove the medium and replace it with fresh Medium I.
NOTE: If the medium becomes yellow on day 4, change the medium on day 4, otherwise on day 5. Add 10 mL of fresh Medium I into the original dish. Hundreds of cysts have already attached to the culture surface; keep them there. - Disperse the cysts in the tube into two 10 cm dishes.
NOTE: Make sure at least 300 cysts are in a dish. If the cysts are not confluent, do not separate the cysts into other dishes; return them to the original 10 cm dish. - On day 7, most cysts (above 95%) attach to the dish, spread out, and form adherent colonies.
NOTE: As early as day 3, the adherent colonies can be observed. - On day 10, change the medium with fresh Medium I. Ensure that all the cysts are attached to the dish.
- Prepare Medium II before the next step by mixing the following: 36 mL of DMEM, 12 mL of F12, 2% of supplement B (v/v), 0.1 mM MEM Non-Essential Amino Acids Solution (NEAA).
- Ensure that the cells are spread out by day 13-17. Perform the next step on day 15 when the cells are spread out but not interacting with the neighboring colonies.
- Rinse the cells once with 1 mL of DPBS and add 1 mL of dispase solution for 5 min at 37 °C.
NOTE: Within 5 min, the edge of the cells elevate. - Remove the dispase buffer and gently rinse the cultures with 1 mL of 1x DPBS.
- Add 10 mL of Medium II to each 10 cm dish and culture in a 37 °C, 5% CO2 incubator.
- The adherent cultures spontaneously detach after 24 h and assemble into retinal organoids.
NOTE: Numerous cells that do not form the organoids would die in the following 2 days. - Three days after detachment, collect the cells from the cell culture dish and allow the organoids to settle in a 15 mL tube.
- Remove the supernatant from the tube and transfer the organoids to a new non-adherent dish (Petri dish) with 10 mL of Medium II for the following four days.
- Prepare Medium III by mixing DMEM: F12 at 3:1 ratio (v/v), 2% supplement B (v/v), 0.1 mM MEM Non-Essential Amino Acids Solution (NEAA), 8% Fetal Bovine Serum (FBS) (v/v), 100 µM of Taurine, and 2 mM Glutamine.
- A week after the detachment, change the medium to Medium III.
- From this day onward, culture the organoids in Medium III and refresh the medium twice a week.
- Establishment of RB cell line.
NOTE: All the reagents are prewarmed at RT.- On day 90, the RBs (80%-90%) encompass the retinal organoids. Therefore, choose the 90-day RB organoids for the generation of the RB cell line.
- Prepare the 1640 medium by mixing the Roswell Park Memorial Institute (RPMI)-1640 medium with 10% FBS.
- Cut the RB into pieces (20-25 pieces per RB) with a microsurgical knife under a microscope in RPMI-1640 medium.
- Remove the RPMI-1640 medium and treat the pieces with 0.25% Trypsin/EDTA for 10 min at 37 °C.
- Add the RPMI-1640 medium to end the reaction and centrifuge at 200 x g for 5 min.
- Remove the supernatant and resuspend the cells in 1640 medium in a non-adherent dish.
- The RB cell line is a floating culture. Change the medium twice a week and passage the RB cells within 2 weeks.
NOTE: The proliferation rate of primary RB cells is quite slow.
Representative Results
The procedure of RB generation is elucidated in the Figure 1, which combines the adherent and floating culture. It was possible to harvest the human RB from RB1-KO hESC, and obtain the RB cell line by isolating the RB organoids.
Here, the protocol provides the details of the differentiation in different stages (Figure 2). Hollow spheres are formed in the first 3 days which attach to the culture surface and then expand (Figure 2A-E). From day 15 onward, cells are elevated and culture in suspension (Figure 2F). The day after the detachment, retinal organoids are formed, and the bright rims are visible (Figure 2G, black arrows). Moreover, those cells outside the organoids are likely to die in the following week (Figure 2G, orange arrows). On day 27, the optic vesicle architecture is evident and around 90% of organoids display this structure (Figure 2H); the organoids without this structure could be discarded. The first detection of the RB occurs on day 45, and then it becomes palpable on day 50 (Figure 2I). When it grows to day 90, the optic vesicle structures are principally enfolded by the RB (Figure 2J). Meanwhile, the RB could be isolated as an RB cell line for further culture (Figure 2K). Above 80% retinal organoids would be fully enveloped by the RB on day 105 (Figure 2L). They highly show expression of Ki67 (proliferation marker) and SYK (oncogene marker) comparing with the H9-derived retinal organoids, which indicates the tumorigenesis in the RB organoids (Figure 2M,N). Additionally, the high expression of ARR3 (cone precursor maker) and CRX (photoreceptor precursor marker) in the RB organoids demonstrates that they originate from cone precursor cells (Figure 2O,P).
The procedure of RB generation mainly undergoes three stages with morphology changes before the RB formation; here, the study provides the inferior and superior results at those stages (Figure 3). Differentiated and undifferentiated hESC (Figure 3A,B) is easy to distinguish from the morphology, and the undifferentiated hESC is chosen for RB formation. On day 5, a hollow sphere should generate (Figure 3D) rather than the solid one (Figure 3C). The RB is derived from the retinal organoids, which display optic vesicle architecture (Figure 3F). There is no RB that would generate in the inferior organoids (Figure 3E).
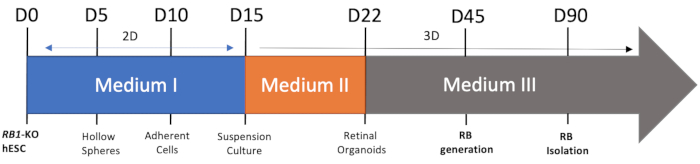
Figure 1: Schematic view of the RB organoids differentiation. Day 0-day 15, the cells are 2D culture in medium I, and after day 15, the cells are suspension culture. RB is formed at around day 45. Please click here to view a larger version of this figure.
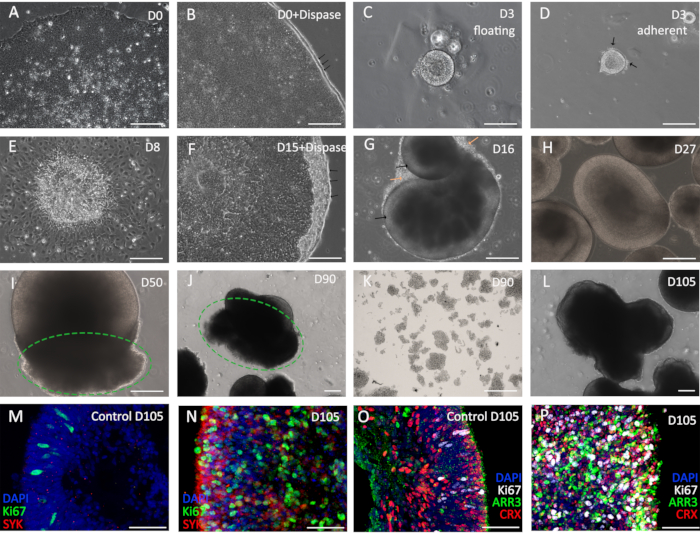
Figure 2: RB generation and characterization. (A–C) The procedure of the early stage, hESC is elevated to form the cysts. The black arrows in (B) show the rolled edges of hESC after dispase treatment. (D,E) The cysts attach to the plates (D) and then expand (E). (F) Adherent cells are elevated to form the retinal organoids. Black arrows indicate the rolled edges after dispase treatment. (G,H) Early days retinal organoids without RB. (I, J) The retinal organoids with RB on day 50 (I) and day 90 (J), the green circles evidence the RB parts. (K) The isolated RB cell line from 90-day retinal organoids. (L) The RB organoids on day 105. (M–P) The immunofluorescence images for the oncogene markers (M,N) and photoreceptor markers (O,P). In A-L, scale bars = 200 µm; in M-P, scale bars = 50 µm. Please click here to view a larger version of this figure.
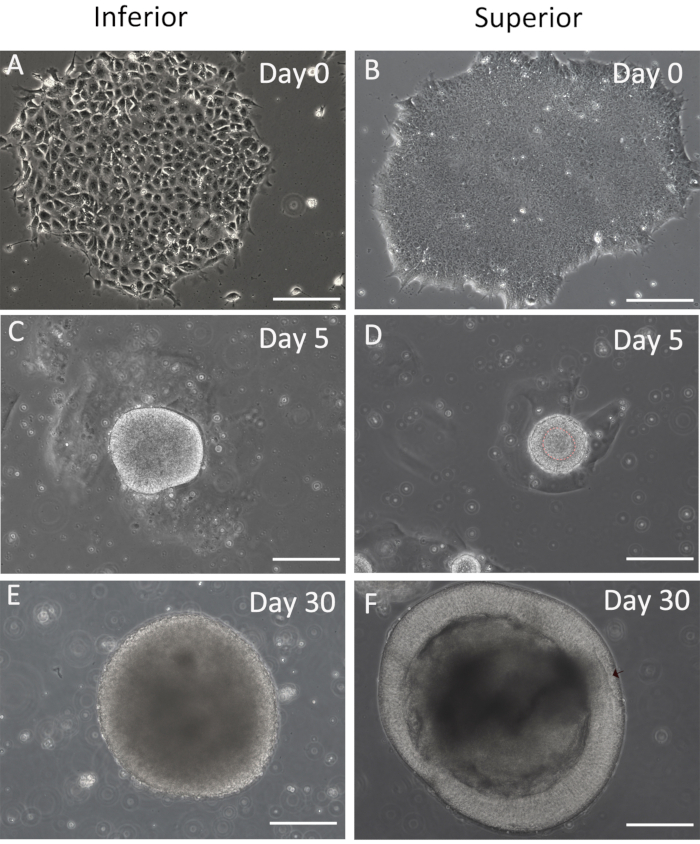
Figure 3: Comparation of the negative and positive results. The inferior and superior images for differentiation on day 0 (A,B), day 5 (C,D), and day 30 (E,F). Scale bars = 200 µm. Please click here to view a larger version of this figure.
Discussion
Human retinoblastoma (RB) is caused by the inactivation of RB1 and the dysfunction of Rb protein. In this protocol, the RB1-KO hESC is the pivotal step for the generation of RB in a dish. While even with RB1-/- hESC, it is possible that there is no RB formation due to the methods of retinal organoid differentiation10. In this protocol, the transfer from adherent culture to floating culture is essential in the process of differentiation. The density of the cysts, types of pluripotent stem cells, and the proliferation rate are all the variables that would affect the timing for detachment. It is desirable to detach the cells when they are expanded but not interacted by neighboring colonies9. If the colonies are adjoining, it would lead to the contiguous retinal organoids and then reduce the differentiation efficiency.
By following the steps critically, it would be untroubled to harvest the RB. However, this method could only model RB with the biallelic inactivation of RB1. For the inherited RB patients, who harbored heterozygous RB1 mutation, it is unable to mimic the process of tumorigenesis with the heterozygous RB1 mutation11. Nevertheless, it is still an optimal RB model because it is currently closest to the actual RB tumorigenesis in patients1. It shares the same origin with primary RB3,12 and overcomes the species difference of mice models or simplified two-dimensional environment of immortalized cancer cell lines3,12,13.
The human RB is established in a dish derived from human ESC using the described method, and it exhibits great similarity to human primary RB. Therefore, it would provide an ideal platform for elucidating the molecular pathology of human RB and screening of pharmacological agents.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thank the 502 team for all the help. This work is partly supported by the Beijing Municipal Natural Science Foundation (Z200014) and National Key R&D Program of China (2017YFA0105300).
Materials
| 2-mercaptoethanol | Life Technologies | 21985-023 | |
| Anti-ARR3 | Sigma | HPA063129 | Antibody |
| Anti-CRX (M02) | Abnove | ABN-H00001406-M02 | Antibody |
| Anti-Ki67 | Abcam | ab15580 | Antibody |
| Anti-Syk (D3Z1E) | Cell Signaling Technology | 13198 | Antibody |
| BbsI | NEB | R3539S | Restriction enzymes |
| Dispase (1U/mL) | Stemcell Technologies | 7923 | |
| DMEM basic | Gibco | 10566-016 | |
| DMEM/F-12-GlutaMAX | Gibco | 10565-042 | |
| DMSO | Sigma | D2650 | |
| DPBS | Gibco | C141905005BT | |
| EDTA | Thermo | 15575020 | |
| Fetal Bovine Serum (FBS), Qualified for Human Embryonic Stem Cells | Biological Industry | 04-002-1A | |
| Glutamine | Gibco | 35050-061 | |
| Ham's F-12 Nutrient Mix (Hams F12) | Gibco | 11765-054 | |
| MEM Non-essential Amino Acid Solution (100X) | Sigma | M7145 | |
| Neurobasal Medium | Gibco | 21103-049 | |
| P3 Primary Cell 4D-Nucleofector X Kit S | Lonza | V4XP-3032 | Nucleofection kit |
| Pen Strep | Gibco | 15140-122 | |
| Puromycin | Gene Operation | ISY1130- 0025MG | |
| QIAquick PCR Purification Kit | QIAGEN | 28104 | |
| ncEpic-hiPSC/hESC culture medium | Nuwacell | RP01001 | ncEpic-hiPSC/hESC culture medium in 1.2.1 |
| Growth factor reduced basement membrane matrix | BD | 356231 | Matrigel in 1.2.1 |
| Cell dissociation enzyme | Gibco | 12563-011 | TrypLE Express in 1.2.8 |
| RNeasy Midi Kit | QIAGEN | 75144 | |
| RNeasy Mini Kit | QIAGEN | 74104 | |
| Supplement A | Life Technologies | 17502-048 | N-2 Supplement (100X), liquid, supplemet in medum I |
| Supplement B | Life Technologies | 17105-041 | B-27 Supplement (50X),liquid, supplemet in medum I,II,III |
| T4 Polynucleotide Kinase | Life Technologies | EK0032 | |
| Taurine | Sigma | T-8691-25G | |
| Y-27632 2HCl | Selleck | S1049 | |
| pX330-U6- Chimeric BB-CBh-hSpCas9-2A-Puro | Addgene | 42230 | |
| Nucleofector 4D | Lonza | ||
| RPMI | Sigma | R0883-500ML |
Riferimenti
- Liu, H., et al. Human embryonic stem cell-derived organoid retinoblastoma reveals a cancerous origin. Proceedings of the National Academy of Sciences of the United States of America. 117 (52), 33628-33638 (2020).
- Singh, H. P., et al. Developmental stage-specific proliferation and retinoblastoma genesis in RB-deficient human but not mouse cone precursors. Proceedings of the National Academy of Sciences of the United States of America. 115 (40), 9391-9400 (2018).
- Xu, X. L., et al. Rb suppresses human cone-precursor-derived retinoblastoma tumours. Nature. 514 (7522), 385-388 (2014).
- Mendoza, P. R., Grossniklaus, H. E. The biology of retinoblastoma. Progress in Molecular Biology and Translational Science. 134, 503-516 (2015).
- Benavente, C. A., Dyer, M. A. Genetics and epigenetics of human retinoblastoma. Annual Review of Pathology. 10, 547-562 (2015).
- Wu, N., et al. A mouse model of MYCN-driven retinoblastoma reveals MYCN-independent tumor reemergence. The Journal of Clinical Investigation. 127 (3), 888-898 (2017).
- Boucherie, C., Sowden, J. C., Ali, R. R. Induced pluripotent stem cell technology for generating photoreceptors. Regenerative Medicine. 6 (4), 469-479 (2011).
- Nakano, T., et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 10 (6), 771-785 (2012).
- Lowe, A., Harris, R., Bhansali, P., Cvekl, A., Liu, W. Intercellular adhesion-dependent cell survival and rock-regulated actomyosin-driven forces mediate self-formation of a retinal organoid. Stem Cell Reports. 6 (5), 743-756 (2016).
- Zheng, C., Schneider, J. W., Hsieh, J. Role of RB1 in human embryonic stem cell-derived retinal organoids. Biologia dello sviluppo. 462 (2), 197-207 (2020).
- Dimaras, H., Corson, T. W. Retinoblastoma, the visible CNS tumor: A review. Journal of Neuroscience Research. 97 (1), 29-44 (2019).
- Xu, X. L., et al. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell. 137 (6), 1018-1031 (2009).
- Qi, D. L., Cobrinik, D. MDM2 but not MDM4 promotes retinoblastoma cell proliferation through p53-independent regulation of MYCN translation. Oncogene. 36 (13), 1760-1769 (2017).

