Generation of Large Numbers of Myeloid Progenitors and Dendritic Cell Precursors from Murine Bone Marrow Using a Novel Cell Sorting Strategy
Summary
Here we provide a method for identifying and isolating large numbers of GM-CSF driven myeloid cells using high speed cell sorting. Five distinct populations (Common myeloid progenitors, granulocyte/macrophage progenitors, monocytes, monocyte-derived macrophages, and monocyte-derived DCs) can be identified based on Ly6C and CD115 expression.
Abstract
Cultures of monocyte-derived dendritic cells (moDC) generated from mouse bone marrow using Granulocyte-Macrophage Colony Stimulating Factor (GM-CSF) have recently been recognized to be more heterogeneous than previously appreciated. These cultures routinely contain moDC as well monocyte-derived macrophages (moMac), and even some less developed cells such as monocytes. The goal of this protocol is to provide a consistent method for identification and separation of the many cell types present in these cultures as they develop, so that their specific functions may be further investigated. The sorting strategy presented here separates cells first into four populations based on expression of Ly6C and CD115, both of which are expressed transiently by cells as they develop in GM-CSF-driven culture. These four populations include Common myeloid progenitors or CMP (Ly6C-, CD115-), granulocyte/macrophage progenitors or GMP (Ly6C+, CD115-), monocytes (Ly6C+, CD115+), and monocyte-derived macrophages or moMac (Ly6C-, CD115+). CD11c is also added to the sorting strategy to distinguish two populations within the Ly6C-, CD115- population: CMP (CD11c-) and moDC (CD11c+). Finally, two populations may be further distinguished within the Ly6C-, CD115+ population based on the level of MHC class II expression. MoMacs express lower levels of MHC class II, while a monocyte-derived DC precursor (moDP) expresses higher MHC class II. This method allows for the reliable isolation of several developmentally distinct populations in numbers sufficient for a variety of functional and developmental analyses. We highlight one such functional readout, the differential responses of these cell types to stimulation with Pathogen-Associated Molecular Patterns (PAMPs).
Introduction
Culturing of murine bone marrow cells with the cytokine Granulocyte-Macrophage Colony Stimulating Factor (GM-CSF) is widely used as a method to generate monocyte-derived dendritic cells (moDC; also known as inflammatory DC) in large numbers 1,2,3,4,5. These cells have been extremely useful in a variety of studies of dendritic cell (DC) function 6,7,8. Typically, these murine bone marrow cells are cultured for 6-8 days and are then used for study of dendritic cell function 5. These cultures had long been considered mostly homogenous, consisting of a majority of differentiated moDC. More recently, it has become clear that at the end of this 6–8 day culture period, there are indeed many moDC, as well as a large subset of differentiated monocyte-derived macrophages (moMacs) 9,10,11. Our own studies have further extended these findings demonstrating that other subsets of less developed cells, such as moDC precursors (moDP) and monocytes, remain in the cultures at low frequency even after 7 days 10. Thus, studies of dendritic cells (DC) function using cells generated by this system could reflect the responses of a broader cohort of cell types than previously appreciated.
We have learned a great deal from the study of GM-CSF-generated moDC relating to the function of these cells in the final stages of differentiation 12,13,14. However, we understand significantly less about the developmental pathway of these cells 2,15,16 and of how and when they exhibit specific functions such as: responsiveness to Pathogen Associated Molecular Patterns (PAMPs), phagocytosis, antigen processing and presentation 13, and anti-bacterial activity. A protocol for isolation of large numbers of conventional Flt3L-driven DC progenitors and precursors has been reported 17. Isolation of these distinct populations was achieved using carboxyfluorescein succinimidyl ester (CFSE)-stained bone marrow cells (to track dividing cells) and culture in Flt3L for 3 days. Cells were then depleted of linage positive cells and sorted into progenitor and precursor populations based on CD11c expression 17. Another approach by Leenen's group to identify early progenitors of DC in GM-CSF-driven culture was to sort cells based on CD31 and Ly6C 18. The initial goal was to create a similar method for obtaining progenitors and precursors of GM-CSF-driven moDC. Due to the specific cell types generated by GM-CSF, we adapted the approach and sorting strategy based on expression of molecules that were expressed at early and later stages of development. We ultimately determined that Ly6C, CD115 (CSF-1 receptor), and CD11c were the best markers for distinguishing these cell types 10.
Here, we present a method for isolation of cells at several distinct stages of development along the pathway of differentiation driven by GM-CSF: Common Myeloid Progenitor (CMP), Granulocyte-Macrophage Progenitor (GMP), monocyte, monocyte-derived Macrophage (MoMac) and monocyte-derived DC (MoDC). The moMac population can be further segregated based on level of MHC class II expression, revealing a moDC precursor population (moDP) 10. We utilize a high-speed fluorescence-activated cell sorting (FACS) strategy to isolate these 5 populations based on expression of Ly6C, CD115, and CD11c. We then demonstrate the examination of these cells in functional assays revealing their responses to PAMP stimulation.
Protocol
All animal work was approved by the Auburn University Institutional Animal Care and Use Committee in accordance with the recommendations outlined in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
1. Preparation for Bone Marrow Collection
- Prepare 250 mL complete media by adding a solution of Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal calf serum, 2 mM glutamine, and 50 µM 2-mercaptoethanol to the top of a 0.22 µm vacuum filter flask unit, and apply vacuum.
NOTE: Complete medium can be stored at 4 °C for up to 2 months. - Prepare 70% ethanol solution by mixing 350 mL of 100% ethanol with 150 mL sterile H2O in a 500 mL flask.
- Set centrifuge to 4 °C.
- Sterilize forceps, scalpel, and scissors in 70% ethanol.
- Using a serological pipette, add 5 mL of complete media to three 60 mm Petri dishes.
- Using a serological pipette, add 30 mL of complete media to a 50 mL conical tube.
2. Collection of Murine Bone Marrow Cells
- Euthanize a C57BL/6 mouse by CO2 narcosis in accordance with the rules established by the 2013 American Veterinary Medical Association (AVMA) Guidelines on Euthanasia.
- Remove and strip hind legs.
- Saturate the hind legs and torso with 75% ethanol, and make shallow cuts through the skin around the hip joint with curved tissue scissors. Using forceps, firmly pull the skin from the hip down towards the ankle, revealing the muscle. Use scissors to remove the skin flap.
- Remove the whole hind leg by cutting through the bone just above the femur/hip joint.
NOTE: In addition to sterilizing the area, the ethanol will aid in providing clean cuts and prevent hair from contaminating the samples. - If the legs will be transferred to a new location for bone marrow harvesting, submerge the legs in complete media.
- Working in a sterile biosafety cabinet, transfer the legs to one of the previously prepared Petri dishes.
- Use scissors to cut the just below the ankle, and carefully remove as much of the muscle and elastic connective tissue as possible. Transfer the cleaned bone to the second prepared Petri dish.
NOTE: Although it is not necessary to remove all the muscle, too much remaining tissue can make it difficult to flush out the bone marrow. - Separate the femur, knee, and tibia.
- Using forceps, hold the leg at the knee and locate the marrow.
NOTE: Bone marrow should be visible as a faint red line inside the bone cavity at the top of the femur and toward the end of the tibia.- Using scissors, make three cuts as follows.
- Cut the tibia just above where the marrow appears to end.
- Cut just below the knee joint.
- Cut just above the knee joint.
- If the hip joint is still connected to the femur, cut just below the hip joint.
- Return the three fragments to the Petri dish and repeat this process on other leg.
- Using scissors, make three cuts as follows.
- Using forceps, hold the leg at the knee and locate the marrow.
- Flush bone marrow from the femur and tibia.
- Fill a 10 mL syringe with complete media from the 50 mL conical tube, and cap with 23 G needle.
- Holding the bone with forceps above the third prepared Petri dish, insert the needle into the bone canal and push media through, flushing out the cells (which may emerge as an intact "plug" or as several clusters). Repeat until no more color can be seen through the bone. Refill the syringe with media as necessary.
- Crush the epiphyses.
- While still in the second Petri dish, hold the knee cap firmly with forceps, and mash the knees with the tip of the syringe. Continue until the epiphyses are no longer red.
- Using the syringe, transfer the cells from the second and third Petri dish to the 50 mL tube. Breakup up clumps by gently pipetting up and down. Try not to generate bubbles. Centrifuge at 250 x g at 4 °C for 10 min.
- Lyse red blood cells
- Remove supernatant with serological pipette, dislodge pellet by flicking, and lyse red blood cells by incubating in 1 mL of ACK (Ammonium-Chloride-Potassium) Lysis Buffer for 1 min at room temperature.
- Using a serological pipette, add 40 mL of HBSS (Hanks Balanced Salt Solution) buffer.
- Using a serological pipette, filter the cells though a 70 µm cell strainer into a new 50 mL conical tube. Centrifuge at 250 x g at 4 °C for 10 min.
- Using a serological pipette, remove supernatant and wash cells with 40 mL of complete media. Centrifuge at 250 x g at 4 °C for 10 min.
NOTE: At this stage, lineage positive lymphocytes can be removed by FACS or magnetic column purification. However, lymphocytes are not maintained long term in culture. Although a large number of lineage positive cells are present in the Ly6C-CD115- population in the bone marrow ex vivo, nearly all are absent by day 5 (Figure 2). - Using a serological pipette, remove the supernatant, and culture the bone marrow cells in complete media with 10 ng/mL of recombinant mouse GM-CSF at a density of 1 x 106 cells/mL.
NOTE: Typically, 4 x 107 total cells can be harvested after red blood cell lysis. However, expect as little as 2 x 107 for beginners and up to 5 x 107 cells for experienced harvesters. - Using a serological pipette, transfer the cells to tissue culture plates, and incubate at 37 °C in 5% CO2. If using a 24-well plate, seed each well with 2 mL of cell suspension.
- Every 48 h, use a serological pipette to remove half of the media and replace with fresh complete media and GM-CSF.
NOTE: Cultures can be kept up to 9 days. However, composition changes over time. See Section 3 for more information.
3. Choosing Day of Sort
- As cell population compositions change over time, select a day that yields the highest number of desired cells.
- See Table 1 for the expected cell yield post sort for each of the populations after 3, 5, and 7 days of culture in GM-CSF per 1 x 107 cells.
4. Staining strategy
- Use small aliquots of cells to prepare control samples. Include an unstained control, compensation control samples stained with only one fluorescent antibody each, and fluorescence-minus-one controls in which all antibodies are added except one, to control for non-specific fluorescence in that channel. If using indirect labeling, include primary alone, secondary alone, and both primary and secondary.
NOTE: Phycoerythrin (PE) and allophycocyanin (APC) tagged antibodies provide distinct separation with minimal bleed over when used together. However, if fluorochrome options are limited, CD115 is expressed at a relatively low level, while Ly6C is expressed at very high levels. Therefore, brighter fluorochromes are desirable for anti-CD115, and anti-Ly6C fluorochromes are chosen to prevent bleed over. - Prepare 100 mL of FACS Wash Buffer (FWB) by mixing 97 mL of chilled Dulbecco's phosphate buffered saline (DPBS) with 3 mL of fetal calf serum in a 50 mL conical tube, and place in an ice bath.
- Use a pipette to gently, but thoroughly, pipette cells up and down to dislodge any loosely adherent cells.
- Using a serological pipette, transfer cells to a 50 mL conical tube. Centrifuge at 250 x g at 4 °C for 10 min.
- If cell volume exceeds 50 mL, transfer cells to the necessary number of tubes and combine at the staining step.
- Gently pour off the supernatant, and wash the pelleted cells by adding 30 mL of FWB with a serological pipette. Centrifuge at 250 x g at 4 °C for 10 min, and repeat the wash.
NOTE: If high cell death is expected, cells can be washed with FWB with as low as 0.5% fetal calf serum (FCS). This will prevent cell clumping. - Suspend and stain cells per antibody manufacturer's instructions.
- Suspend 5 x 107 cells in 1 mL of FWB and add 2 µg each of anti-Ly6C and anti-CD115 labeled with the fluorophores (of the researcher's choice). To further distinguish CMP from moDC (both are Ly6C- and CD115-), add 2 µg of anti-CD11c antibodies (CMP are CD11c–; moDC are CD11c+). Incubate for 30 min on ice.
NOTE: Figure 3 was generated using Ly6C-PE and CD115-APC.
- Suspend 5 x 107 cells in 1 mL of FWB and add 2 µg each of anti-Ly6C and anti-CD115 labeled with the fluorophores (of the researcher's choice). To further distinguish CMP from moDC (both are Ly6C- and CD115-), add 2 µg of anti-CD11c antibodies (CMP are CD11c–; moDC are CD11c+). Incubate for 30 min on ice.
- Using a serological pipette, add 10 mL of FWB, and centrifuge at 250 x g at 4 °C for 10 min.
- Gently pour off the supernatant, and wash the pelleted cells by adding 30 mL of FWB with a serological pipette. Centrifuge at 250 x g at 4 °C for 10 min, and repeat the wash.
- Before suspending cells, flick tube thoroughly to dislodge the pellet. Use a serological pipette to suspend cells at 1 x 107 cell/mL of FWB, and filter through 35-µm cell filter. Use a serological pipette to transfer filtered cells into polypropylene tube, and place on ice until ready to sort.
NOTE: If polypropylene is unavailable, protein coated (nonfat dry milk or FCS) polystyrene tubes can be used to reduce binding.
5. Set Gates Based on Control Samples
NOTE: To prevent cell disruption due to the pressure of the high-speed flow stream, use a 100–130 µm nozzle for cell sorting.
- Run the unstained control (see step 4.1) through the cell sorter, and apply a gate to exclude small debris (low forward scatter; FSC) and highly granular (high side scatter; SSC) particles.
- To analyze only the later stages (monocytes, moMac/MoDP, and MoDC), apply gating to only include larger cells (high FSC).
- If viability stains are being used, use these to exclude stained, non-viable events (an example is illustrated in Figure 1).
- Run the single fluorescent control samples through the cell sorter, and adjust compensation as needed.
- Run a sample of the multi-labeled sample. Observe four distinct populations: Ly6C+CD115- (GMPs), Ly6C+CD115+ (monocytes), Ly6C-CD115+ (moMacs/moDP), and Ly6C-CD115- (CMPs/moDCs). Apply a gate to isolate each of the four major populations.
NOTE: CMPs and moDC share the Ly6C-CD115- phenotype. However, they can be differentiated based on CD11c expression: CMP lack CD11c, whereas MoDCs express CD11c.
6. Collection of Isolated Populations
- Prepare collection tubes by adding enough FCS to achieve at least 20% final concentration when full. For example, if using 5 mL tubes, add 1 mL of FCS before sorting, and remove the tube when it reaches 5 mL total volume.
- To prevent membrane turnover and antibody uptake, keep all samples (mixed and sorted) at 4 °C throughout the sort.
- If this is not possible, keep the tube containing the cells to be sorted on ice as much as possible and transfer aliquots to the sorter as needed.
- Additionally, transfer sorted samples to ice every 20–30 min.
- After the desired number of cells have been collected, use a serological pipette to transfer the cells to a new conical tube. Centrifuge at 250 x g at 4 °C for 10 min.
- Confirm purity with post-sort analysis on small aliquots from each collected population.
- Remove the supernatant, suspend in 10 mL of FWB and centrifuge at 250 x g at 4 °C for 10 min. Repeat for a total of two washes.
NOTE: It can be difficult to remove all the supernatant without dislodging the pellet. Attaching a pipette tip to a vacuum line can help with removal. If this is not available, it is suggested that the supernatant be collected in a fresh tube in case of pellet dissociation. - Remove the supernatant after the second wash.
- If the user's experimental design dictates the cells be re-cultured, follow steps 2.12–2.14. Otherwise, if cells will be used for immediate analysis, prepare cells according to desired protocol.
NOTE: An example of typical functional analysis is included in Figure 4.
Representative Results
In an effort to keep as many channels available for analysis as possible, viable cells were routinely selected based on forward and side scatter, excluding very small and very granular events (a typical gate is applied to all the dot plots in Figure 1A). To determine if this gating strategy reliably excluded dead cells, we stained with 7-Amino actinomycin D (7-AAD) (Figure 1B). 7AAD stains DNA in dead and dying cells due to membrane permeability, and is excluded by viable cells. Viability of GM-CSF cultured bone marrow cells was analyzed immediately post-harvest (PH) and 1, 3, and 5 PH. Cell analyzed immediately PH had approximately 10% of 7-AAD positive cells when a typical FSC/SSC gate was applied to freshly isolated cells from the bone marrow (Figure 1, Post-Harvest). A similar proportion of dead cells was also present at day 1 (~12%) and day 3 (~11%) of culture (Figure 1, 1 day PH and 3 days PH). By day 5, the number of dead cells within the gate was reduced to ~5% (Figure 1, 5 days PH). Thus, using such a viability gate is generally appropriate for sorting on day 5 and after. Therefore, if users are limited in their available parameters, FSC/SSC gating is generally appropriate. However, for more sensitive assays (particularly if they rely on precise cell numbers), incorporate a viability stain (suggestion).
Many protocols for propagation of dendritic cells recommend depletion of lineage positive cells, especially lymphocytes, from bone marrow prior to culture 11,17. This procedure is thought to increase the purity of the cells recovered upon GM-CSF-mediated differentiation. Typically, cells expressing markers of T cells (CD3), B cells (CD45R or CD19), and NK cells (NK1.1) may be depleted by positive selection using magnetic beads or cell sorting 11,17,18. However, based on the culturing system, lymphocytes were rarely recovered or detected in these assays. Thus, we sought to assess the longevity of lymphocytes maintained in the Ly6C-CD115- population among the GM-CSF-driven cells (Figure 2A). GM-CSF cultured bone marrow cells (that had not been lineage depleted) were stained with antibodies to CD3, CD45R, and NK1.1 (in the same fluorophore) and measured daily by flow cytometry (Figure 2B). Within the Ly6C-CD115- population, CD3/CD45R positive cells persisted strongly through Days 0–3 (Figure 2B). On Day 4, only a few CD3/CD45R positive cells remained, and by Day 5 and 6, there were no CD3/CD45R expressing cells present. Thus, within 4 days of culture in GM-CSF, lineage positive cells were essentially absent and were not detected at all at days 5 and 6 of culture.
The composition of the GM-CSF-driven cell culture changes daily in this system as the cells develop and differentiate (Table 1; Figure 3). At early time points, the most abundant cells are progenitors and precursors, and at later times the majority of cells are more differentiated 10. To determine how sorting on different days of culture might affect the subsequent developmental path or kinetics, sorts were performed 3, 5, and 7 days PH (Figure 3A). The development of the MoMac population (Ly6C-CD115+) was then tracked over 2–3 further days in culture (Figure 3B-3D).
When sorted 3 days PH, only 40% of Ly6C-CD115+ cells had decreased CD115 expression within 24 h post-sort (PS) (Figure 3B). By 48 h PS, the fraction that had down-regulated CD115 was 66%, and by 72 h, 70% of the cells had this phenotype. This phenotypic composition was maintained (~70-72% Ly6C-CD115-) even after further days of culture (data not shown). When sorted 5 days PH, ~75% of the cells were Ly6C-CD115-, having rapidly down-regulated CD115 within 24 h PS, and ~80% were CD115- after only 48 h. This distribution was maintained after 72 h (Figure 3C). Finally, when sorted 7 days PH, down-regulation of CD115 was also quite rapid. Within 24 h PS, ~75% of cells had down-regulated CD115 expression, and this trend was maintained after 48 h (Figure 3D). Interestingly, when sorted at day 7, the overall level of CD115 expression was lower on the cells within the CD115+ population.
Thus, these findings indicate that the kinetics of development are somewhat slower when sorting at an early day, such as day 3 sorted cells compared to the more rapid development and differentiation observed after sorting on day 5 or 7. Based on these results, a user seeking larger numbers of moDC should likely sort on day 5 or 7.
The maturation response of DC is well established 6,7,12,14. When treated with a variety of pathogen-associated molecular patterns (PAMPs), immature DC up-regulate the expression of MHC, costimulatory molecules, and pro-inflammatory cytokines, enhancing their T cell-activating capacity 6. However, it is less clear when developing cells gain the capacity to respond to PAMP stimulation and which feature of DC maturation they might exhibit. To determine which of the sorted populations would respond to maturation stimuli, each population was treated shortly after sorting with a cocktail of PAMPs including Poly I:C, lipopolysaccharides (LPS), and CpG DNA to trigger toll-like receptor (TLR) 3 (TLR3), TLR4, and TLR9, respectively. Cells were treated (or untreated) for 24 h and expression of CD86 and MHCII was measured by flow cytometry (Figure 4A–4B). Furthermore, IL-12p40/70 and IL-6 production was measured in the supernatants by cytokine array ELISA (Figure 4C-4D).
CMPs and GMPs expressed very low levels of MHC class II and CD86 in an unstimulated state, and these expression patterns did not change significantly upon exposure to the cocktail of PAMPs (Figure 4A and 4B). Likewise, the expression of MHC class II by the monocytes was also low and changed little upon PAMP exposure (Figure 4A). However, the expression of CD86 was moderate on the monocytes 24 h after sorting, and it increased further following 24 h stimulation. Higher basal expression of MHC class II and CD86 in the MoMacs and MoDCs was observed, and both populations exhibited a strong induction of these molecules upon PAMP stimulation. In terms of MHC class II expression following stimulation, there was no statistical difference between the CMPs, GMPs, and monocytes, while the MoMacs and MoDCs formed a distinct group. Yet, in terms of CD86 expression, the CMPs and GMPs cells were statistically different than MoMacs and MoDCs. However, monocytes were not different than MoMacs or MoDCs.
We next wanted to assess the relative ability of each of the five populations to produce cytokines in response to TLR stimulation. Thus, we performed a cytokine assay on the sorted populations after culture in the presence or absence of the TLR agonist cocktail used above. Cells were cultured with the stimuli for 24 h, then supernatants were collected. We observed very little IL-12p40/70 or IL-6 production by CMPs upon TLR stimulation (Figure 4C-4D). The second population, GMPs, were unable to produce IL-12p40/70 (Figure 4C), but these cells produced a low amount of IL-6 upon stimulation by PAMPs (Figure 4D). The highest levels of cytokine production were observed in the latter three populations. Monocytes and MoMacs had very similar patterns and magnitudes of cytokine production. Both greatly increased IL-12p40/70 and modestly increased IL-6 production up stimulation (Figure 4C-4D). Interestingly, in the presence of PAMPs MoDCs did not increase IL-12p40/70 secretion; however, this population greatly increased IL-6 production (Figure 4C-4D). These findings indicate that the sorted populations maintained their immunological functions after isolation.
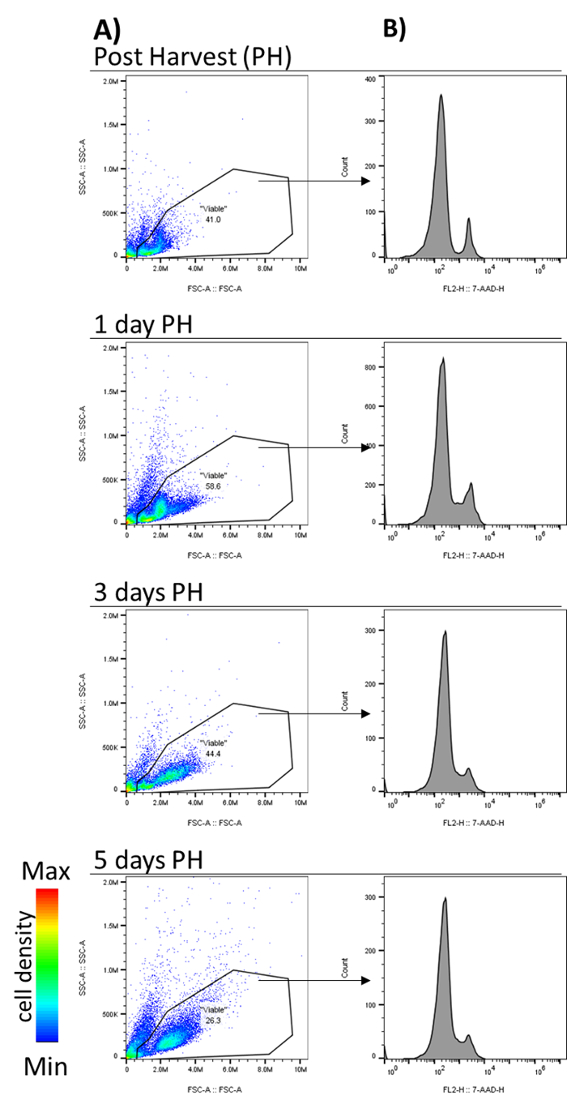
Figure 1: Viable cells within Forward versus Side Scatter gate. Mouse bone marrow was cultured in GM-CSF, and viability was measured using 7-AAD (7-amino-actinomycin D) staining post-harvest (PH) and 1, 3, and 5 days post-harvest. (A) Viable cells were selected by applying a gate (Viable) which omitted small and highly granular events based on FSC and SSC. (B) Histograms of 7-AAD staining generated from events within Viable (FSC/SSC) gate. Events positive for 7-AAD indicate cells undergoing apoptosis. Arrows indicate viable cell gating. Please click here to view a larger version of this figure.
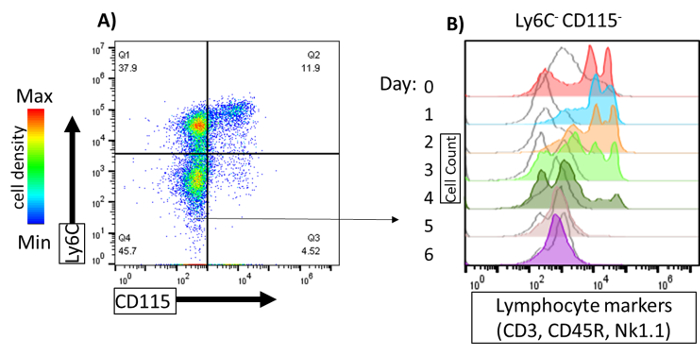
Figure 2: Lymphocytes within the Ly6C-CD115- population. Mouse bone marrow was cultured in GM-CSF and lymphocyte markers (CD3, CD145R, and NK1.1) were analyzed daily by flow cytometry. (A) Cell were stained with Ly6C-PE and CD115-APC to identify Ly6C-CD115- cells. Quadrant gate was applied based on single-color controls. Pseudocolor dot plot generated from day 0 (B) CD3, CD45R, and NK1.1 expression of Ly6C-CD115- cells was analyzed on day of harvest (day 0) until day 6. Cell counts were normalized to mode using a flow cytometry analysis software. Arrow indicates Ly6C-CD115- gating. Please click here to view a larger version of this figure.
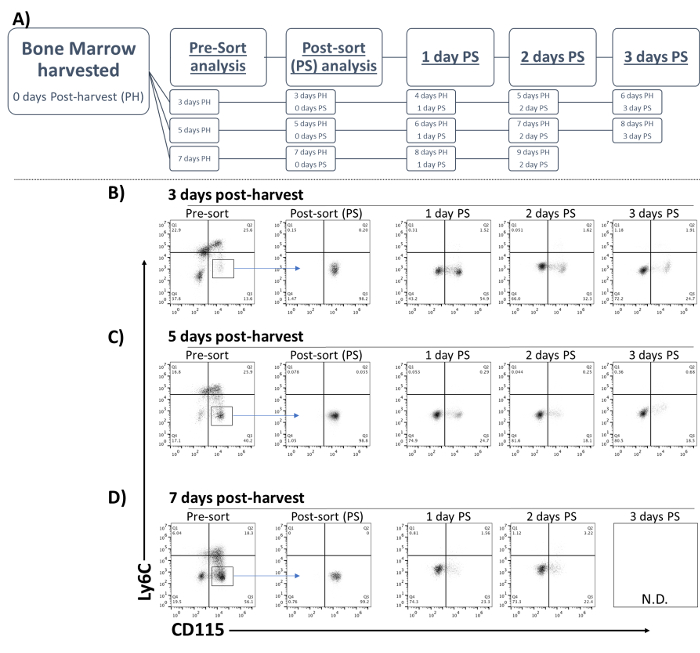
Figure 3: Kinetics of development of Ly6C-CD115+ cells after sorting on different days. (A) Mouse bone marrow was harvested and cultured in GM-CSF. Aliquots of 1 x 107 cells were harvested (B) 3, (C) 5, or (D) 7 days post-harvest (PH). On the indicated days PH, Ly6C-CD115+ cells were sorted from mixed culture (Pre-sort) and analyzed immediately post-sort (PS). Sorted cells were re-cultured in GM-CSF, and changes in Ly6C/CD115 expression were analyzed daily by flow cytometry. Box and arrow indicate sorting gate. Please click here to view a larger version of this figure.
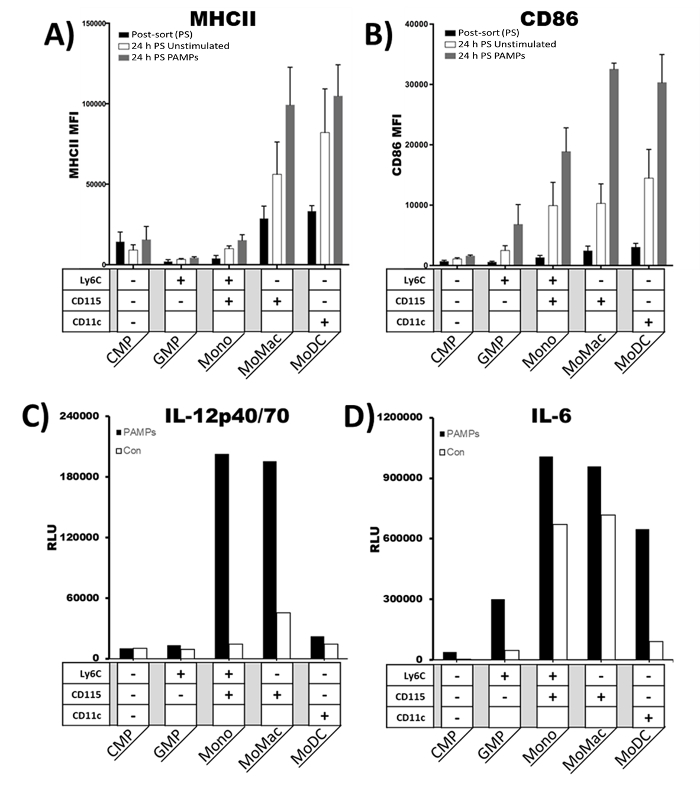
Figure 4: Maturation and cytokine expression following TLR stimulation. Cells were sorted into 5 populations on day 3 of culture in GM-CSF. They were then treated with a cocktail of PAMPs (LPS, Poly I:C, and CpG DNA) for 24 h. Mean fluorescent intensity (MFI) of (A) MHC class II and (B) CD86 was analyzed immediately post-sort (PS) and 24 h with and without PAMPs by flow cytometry. Error bars represent standard deviation of 3-5 replicate experiments. (C) IL-12p40/70 and (D) IL-6 in supernatants from 3 pooled samples were measured by cytokine array dot blot ELISA after 24 h with and without PAMPs. RLU; Relative light units; -/+ indicate the presence of the cell surface marker for the designated population. Please click here to view a larger version of this figure.
| Day 3 | Day 5 | Day 7 | |||||
| Phenotype | Cell type | Min | Max | Min | Max | Min | Max |
| Ly6C-CD115-CD11c- | CMP | 3 x 106 | 4 x 106 | 5 x 105 | 1 x 106 | N/a | N/a |
| Ly6C+CD115- | GMP | 2 x 106 | 3 x 106 | 2 x 106 | 3 x 106 | N/a | N/a |
| Ly6C+CD115+ | Mono | 2 x 106 | 3 x 106 | 2 x 106 | 3 x 106 | 5 x 105 | 1 x 106 |
| Ly6C-CD115+ | MoMac | 5 x 105 | 1 x 106 | 3 x 106 | 4 x 106 | 3 x 106 | 4 x 106 |
| Ly6C-CD115-CD11c+ | MoDC | N/a | N/a | 5 x 105 | 1 x 106 | 3 x 106 | 4 x 106 |
Table 1: Expected minimum and maximum number of cells recovered post-sort per 1 x 107 cells. CMP (common myeloid progenitor); GMP (granulocyte/macrophage progenitor) Mono (monocytes); MoMac (monocyte-derived macrophage); MoDC (monocyte-derived dendritic cell); N/a (not available); Min (minimum cell yield out of 1 x 107 cells); Max (maximum cell yield out of 1 x 107 cells).
Discussion
This protocol facilitates isolation of GM-CSF-driven progenitor and precursor cell types in numbers sufficient for several types of analyses including biochemical assays, assays of cellular function in vitro, or instillation in vivo. This method represents a significant advance in the field of monocyte-derived dendritic cell development, enabling the reliable isolation and identification of cells early in this pathway of development as well as those differentiated cell types more commonly isolated in prior studies.
Previous protocols for isolation of progenitors and precursors generated from bone marrow in vitro have relied on CFSE-staining to identify proliferative progenitor cells 17 or on staining with CD31 and Ly6C as markers of myeloid cells 18. The CFSE staining protocol described by Naik was designed for use in generation of Flt3L-driven DC progenitors and precursors 17. When we attempted this approach in the GM-CSF culture system, we encountered two main issues. The CFSE was somewhat cytotoxic to the developing cells and was so bright (even in divided cells) that it made compensation difficult. This approach proved to be unsuitable for our goals of isolating early cell types (progenitors), as well as cells across the developmental spectrum, from highly proliferative (CMPs/GMPs) to highly developed (MoDC/MoMac). We also tried the sorting strategy for GM-CSF-driven cells described by Leenen's group which was based on CD31 and Ly6C 18. However, we found that CD31 was problematic. It was expressed at very low levels (making it difficult to resolve the populations for clean sorting) by a vanishingly small subset of cells, and only very early during the culture period. In fact, CD31 expression was not detectable past day 2 of culture in GM-CSF 10.
Ly6C, however, was a very useful molecule for separating cells at different stages of development due to its transient pattern of expression 10. The addition of CD115 enabled us to more closely discern cells at intermediate and later stages of development that was not possible with CD31. We also examined other markers of progenitors such as CD34 and CD117 in hopes of isolating large numbers of these early cells 10. However, unlike that reported in the Flt3L system, we found that CD34 and CD117 were also expressed on a very small subset of cells and were virtually absent by day 3 of culture 17. These stem cell markers may be useful in future studies however, to distinguish subsets within the CMP or GMP populations.
There are several potential modifications to the protocol that may affect the yield of desired cell populations. First, the user may choose to apply a viability stain to exclude any dead cells. Based on the observations, the rate of dead cells within a typical forward and side scatter gate are relatively low in general, and pose problems only during the first few days of culture, coincident with decreases in lineage positive lymphocytes. However, for applications in which cell numbers must be precise, a viability stain will ensure exclusion of dead cells.
A second potential modification is depletion of lineage-positive lymphocytes prior to culture. We have tried this approach to address the small population of lineage positive cells that were regularly observed within the Ly6C-CD115- cell population. The overall cell yield was slightly lower at days 5 and 6, and the purity was not significantly higher (data not shown). Thus, the purity did not justify the reduced yield. Likewise, if the user plans to sort on day 5 or after, the frequency of lymphocytes within the culture is well below 1% and should not present a problem.
Finally, because this strategy is designed to isolate cells across a large developmental spectrum the user should tailor the timing of their sort to collect as many of the desired populations as possible. This sorting strategy faithfully yields the cell types indicated regardless of the day of culture on which the cells are sorted. For example, cells with the phenotype Ly6C+ CD115- CD11c- are true to the GMP phenotype and function similarly whether they are sorted and isolated on day 3 or 5 of culture. However, the frequency of these cells is much greater on day 3 than 5, so if the goal is isolation of this cell type, sorting on day 3 would be recommended. It is also notable that cells sorted on day 3 progress through the subsequent developmental stages with slower kinetics than if they were sorted on day 5 or 7 (Figure 3).
While this method allows for the clear delineation and isolation of 5–6 distinct populations along the developmental spectrum, there are likely many more populations or transitional stages along this pathway. Within each of the five described populations there may be several sub-populations at slightly different increments of development. For example, we have observed "distinct" populations with intermediate levels of CD115 and of Ly6C which undergo slightly different patterns of development, in terms of how many moMac and moDC are generated (data not shown). It is also likely that populations previously observed in vivo are present in the culture and sorting system, but these have been difficult to identify due to their very low frequency. Populations such as cMOP 19, MDP 20, or CDP 21 are likely present, but may be obscured within the larger populations. Future studies will be needed to further clarify the numerous developmental stages that may be isolated within this sorting framework with the addition of specific markers of differentiation. The value of this sorting strategy is that it allows consistent isolation of large numbers of developmentally distinct stages during DC ontogeny.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We are grateful for technical assistance from Alison Church Bird at the Auburn University School of Veterinary Medicine Flow Cytometry Facility, for funding from the NIH to EHS R15 R15 AI107773 and to the Cellular and Molecular Biology Program at Auburn University for summer research funding to PBR.
Materials
| RPMI 1640 | Corning | 15-040-CV | |
| Fetal Calf Serum (FCS) | HyClone | SV30014.04 | to supplement complete medium and FWB |
| GlutaMAX | Gibco | 35050 | to supplement complete medium |
| 2-mercaptoethanol (2-ME) | MP Biomedical | 190242 | to supplement complete medium |
| 75mM Vacuum Filter | Thermo Scientific | 156-4045 | to sterilize complete media |
| ACK Lysis Buffer | Lonza | 10-548E | to lyse red blood cell |
| HEPES buffer | Corning | 21-020-CM | to rescue leukocytes after red blood cell lysis |
| Phosphate Buffered Saline (PBS), Dulbecco's | Lonza | 17-512F | must be endotoxin free; chilled at 4 °C |
| 35µm Cell filter | Falcon | 352235 | to break apart clumps before running through cytometer. |
| GM-CSF | Biosource | PMC2011 | usable concentration of 10ng/mL |
| Tissue cultured treated plate | VWR | 10062-896 | for bone marrow cells after harvest |
| Anti-Ly6C, Clone HK1.4 | Biolegend | 128018 | |
| Anti-CD115, Clone AFS98 | Tonbo Bioscience | 20-1152-U100 | |
| Anti-CD11c, Clone HL3 | BD Biosciences | 557400 | to differeniate CMP and MoDCs |
| MoFlo XPD Flow Cytometer | Beckman Coulter | ML99030 | |
| BD Accuri C6 | BD Biosciences | 660517 | |
| 100% Ethanol | Pharmco-Aaper | 111000200CSPP | |
| 60mm Petri Dish | Corning, Inc | 353002 | |
| 50mL Conical tube | VWR | 21008-242 | |
| C57BL/6 Mice | The Jackson Laboratory | 000664 | Female; 10-20 weeks old |
| Biosafety Hood | Thermo Scientific | 8354-30-0011 | |
| 10mL Syringe | BD Biosciences | 301604 | |
| 23-gauge needle | BD Biosciences | 305145 | |
| Centrifuge 5810 R | eppendorf | 22625501 | |
| FlowJo Software v10 | BD Biosciences | Version 10 | flowjo.com |
References
- Zhan, Y., Xu, Y., Lew, A. M. The regulation of the development and function of dendritic cell subsets by GM-CSF: more than a hematopoietic growth factor. Mol Immunol. 52, 30-37 (2012).
- Zhan, Y., et al. The inflammatory cytokine, GM-CSF, alters the developmental outcome of murine dendritic cells. Eur J Immunol. , (2012).
- van de Laar, L., Coffer, P. J., Woltman, A. M. Regulation of dendritic cell development by GM-CSF: molecular control and implications for immune homeostasis and therapy. Blood. 119, 3383-3393 (2012).
- Steinman, R. M., Inaba, K. Myeloid dendritic cells. J Leukoc Biol. 66, 205-208 (1999).
- Inaba, K., et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 176, 1693-1702 (1992).
- Steinman, R. M., Pack, M., Inaba, K. Dendritic cell development and maturation. Adv Exp Med Biol. , 1-6 (1997).
- Pierre, P., et al. Developmental regulation of MHC class II transport in mouse dendritic cells. Nature. 388, 787-792 (1997).
- Steinman, R. M., Witmer-Pack, M., Inaba, K. Dendritic cells: antigen presentation, accessory function and clinical relevance. Adv Exp Med Biol. , 1-9 (1993).
- Na, Y. R., Jung, D., Gu, G. J., Seok, S. H. GM-CSF Grown Bone Marrow Derived Cells Are Composed of Phenotypically Different Dendritic Cells and Macrophages. Mol Cells. 39, 734-741 (2016).
- Rogers, P. B., Driessnack, M. G., Hiltbold Schwartz, E. Analysis of the developmental stages, kinetics, and phenotypes exhibited by myeloid cells driven by GM-CSF in vitro. PLoS One. 12, e0181985 (2017).
- Helft, J., et al. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCII(+) Macrophages and Dendritic Cells. Immunity. 42, 1197-1211 (2015).
- Alloatti, A., Kotsias, F., Magalhaes, J. G., Amigorena, S. Dendritic cell maturation and cross-presentation: timing matters!. Immunol Rev. 272, 97-108 (2016).
- Jakubzick, C. V., Randolph, G. J., Henson, P. M. Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol. 17, 349-362 (2017).
- Mbongue, J. C., Nieves, H. A., Torrez, T. W., Langridge, W. H. The Role of Dendritic Cell Maturation in the Induction of Insulin-Dependent Diabetes Mellitus. Frontiers in immunology. 8, 327 (2017).
- Shortman, K., Naik, S. H. Steady-state and inflammatory dendritic-cell development. Nat Rev Immunol. 7, 19-30 (2007).
- Xu, Y., Zhan, Y., Lew, A. M., Naik, S. H., Kershaw, M. H. Differential development of murine dendritic cells by GM-CSF versus Flt3 ligand has implications for inflammation and trafficking. J Immunol. 179, 7577-7584 (2007).
- Naik, S. H. Generation of large numbers of pro-DCs and pre-DCs in vitro. Methods Mol Biol. 595, 177-186 (2010).
- Nikolic, T., de Bruijn, M. F., Lutz, M. B., Leenen, P. J. Developmental stages of myeloid dendritic cells in mouse bone marrow. Int Immunol. 15, 515-524 (2003).
- Hettinger, J., et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol. 14, 821-830 (2013).
- Fogg, D. K., et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 311, 83-87 (2006).
- Traver, D., et al. Development of CD8{alpha}-Positive Dendritic Cells from a Common Myeloid Progenitor. Science. 290, 2152-2154 (2000).

