Submillisecond Conformational Changes in Proteins Resolved by Photothermal Beam Deflection
Summary
Here we report an application of the photothermal beam deflection technique in combination with a caged calcium compound, DM-nitrophen, to monitor microsecond and millisecond dynamics and energetics of structural changes associated with the calcium association to a neuronal calcium sensor, Downstream Regulatory Element Antagonist Modulator.
Abstract
Photothermal beam deflection together with photo-acoustic calorimetry and thermal grating belongs to the family of photothermal methods that monitor the time-profile volume and enthalpy changes of light induced conformational changes in proteins on microsecond to millisecond time-scales that are not accessible using traditional stop-flow instruments. In addition, since overall changes in volume and/or enthalpy are probed, these techniques can be applied to proteins and other biomacromolecules that lack a fluorophore and or a chromophore label. To monitor dynamics and energetics of structural changes associated with Ca2+ binding to calcium transducers, such neuronal calcium sensors, a caged calcium compound, DM-nitrophen, is employed to photo-trigger a fast (τ < 20 μsec) increase in free calcium concentration and the associated volume and enthalpy changes are probed using photothermal beam deflection technique.
Introduction
Photo-thermal methods such as photoacoustic calorimetry, photothermal beam deflection (PDB), and transient grating coupled with nanosecond laser excitation represent a powerful alternative to transient optical spectroscopies for time-resolved studies of short-lived intermediates1,2. In contrast to optical techniques, such as transient absorption and IR spectroscopy, that monitor the time profile of absorption changes in the chromophore surrounding; photothermal techniques detect the time-dependence of heat/volume changes and therefore are valuable tools for investigating time profiles of optically “silent” processes. So far, photoacoustic calorimetry and transient grating has been successfully applied to study conformational dynamics of photo-induced processes including diatomic ligand migration in globins3,4, ligand interactions with oxygen sensor protein FixL5, electron and proton transport in heme-copper oxidases6 and photosystem II as well as photo-isomerization in rhodopsin7 and conformational dynamics in cryptochrome8.
To expand the application of photothermal techniques to biological systems that are lacking an internal chromophore and/or fluorophore, the PBD technique was combined with the use of caged compound to photo-trigger an increase in ligand/substrate concentration within few microseconds or faster, depending on the caged compound. This approach allows monitoring of dynamics and energetics of structural changes associated with the ligand/substrate binding to proteins that are lacking an internal fluorophore or chromophore and on time-scale that are not accessible by commercial stop-flow instruments. Here an application of PBD to monitor the thermodynamics of the cage compound, Ca2+DM-nitrophen, photo-cleavage as well as the kinetics for Ca2+ association to the C-terminal domain of the neuronal calcium sensor Downstream Regulatory Element Antagonist Modulator (DREAM) is presented. The Ca2+ is photo-released from Ca2+DM-nitrophen within 10 μsec and rebinds to an unphotolysed cage with a time constant of ~300 μsec. On the other hand, in the presence of apoDREAM an additional kinetic occurring on the millisecond time-scale is observed and reflects the ligand binding to the protein. The application of PBD to probe conformational transitions in biological systems has been somehow limited due the instrumental difficulties; e.g. arduous alignment of the probe and pump beam to achieve a strong and reproducible PBD signal. However, a meticulous design of an instrumentation set-up, a precise control of the temperature, and a careful alignment of the probe and pump beam provide a consistent and robust PBD signal that allows monitoring of time-resolved volume and enthalpy changes on a broad time-scale from 10 µsec to approximately 200 msec. In addition, modifications of the experimental procedure to assure the detection of sample and reference traces under identical temperature, buffer composition, optical cell orientation, laser power, etc. significantly reduces the experimental error in measured reaction volumes and enthalpies.
Protocol
1. Sample Preparations
- Carry out the sample preparation and all sample manipulations in a dark room to prevent an unwanted uncaging.
- Solubilize DM-nitrophen ((1-(2-nitro-4,5-dimethoxyphenyl)-N,N,N’,N’-tetrakis [(oxy-carbonyl) methyl]-1,2-ethanediamine) in 50 mM HEPES buffer, 100 mM KCl, pH 7.0 to a final concentration of 400 µM (ε350nm = 4330 M-1cm-1 9).
- Add CaCl2 from the 0.1 M stock solution to achieve a desirable ratio of [Ca2+]:[DM-nitrophen]. For proteins with Kd for Ca2+ association larger than 10 µM, the ratio of [Ca2+]:[DM-nitrophen] of 1:1 is preferable to prevent binding of photo-released Ca2+ to uncaged DM-nitrophen. Indeed, considering the Kd value for DM-nitrophen to be 10 nM and the total concentration of DM-nitrophen and Ca2+ to be 400 mM, ~90% of a calcium binding protein with Kd = 10 μM will be in the apoform. On the other hand, for study of Ca2+ binding to proteins with Kd < 10 µM, it’s preferable to decrease the [Ca2+]:[DM-nitrophen] ratio to 0.95 to prevent Ca2+ complexation with the apo-protein prior the cage photo-dissociation.
- Solubilize the reference compound, K3[Fe(III)(CN)6] or Na2CrO4, in the same buffer as for the sample.
2. Setting up the Experiment
- The basic experimental configuration is shown in Figure 2.
- Use a pin hole (P2) to adjust the diameter of the probe beam (632 nm output of He-Ne laser, ~5 mW laser power) to 1 mm and propagate the probe beam through the center of a cell placed in a temperature controlled cell holder using a M1 mirror.
- Use a mirror (M2) behind the sample to center the probe beam on a center of position sensitive detector.
- Focus the probe beam on the center of the detector in such way that the difference in the voltage between the top two diodes and bottom two diodes as well as the difference in the voltage between the two diodes on the left and right side of the detector is zero.
- Subsequently, shape a diameter of the pump beam, a 355 nm output of Q-switched Nd:YAG laser, FWHM 5 nsec) using a 3 mm pinhole (P1) placed between two 355 nm laser mirrors.
- Copropagate the pump beam through the center of the cuvette as demonstrated in Figure 2. It is important that both laser beams are propagated through the center of the optical cell in nearly colinear manner to obtain a measurable deflection angle and thus high amplitude of PBD signal. Under experimental conditions, the angle of the intersection of the probe and pump beams is less than 15°.
- Use a reference compound to align the probe and pump beam to achieve a satisfactory PBD signal, i.e. a good S/N ratio and stable PBD amplitude on longer time-scales (~100 msec).
- Adjust the position of the pump beam with respect to the probe beam by an incremental adjustment of 355 nm laser mirrors.
- Measure the amplitude of the PBD reference signal as a difference in between two top and bottom photodiodes on the position sensitive detector. The PBD signal should exhibit a rapid increase in the amplitude on a fast time-scale (<10 µsec) and remain stable on 100 msec timescale as demonstrated in Figure 3. The shot to shot variability of the PDB amplitude is within 5% of the signal amplitude and the signal reproducibility is mainly affected by vibrations.
- Check the linearity in the PBD signal amplitude with respect to the released heat energy by measuring the linear dependence of the PBD signal on the excitation laser power and on the number of Einsteins absorbed, Ea = (1-10-A), where A corresponds to the reference absorbance at the excitation wavelength.
- Keep the laser power below approximately 1,000 µJ and absorbance of the sample/reference compound at the excitation wavelength less than 0.5 to prevent multiphoton absorption and decrease of the pump beam power, respectively, and ensure linearity of the PBD signal.
3. PBD Measurements
- Start with the measurement of the PBD traces for the reference. Place the solution of the reference compound in a 1.0 cm x 1.0 cm or 1.0 cm x 0.5 cm quartz cell and position the cell in the temperature controlled holder. Both path lengths provide comparable PBD amplitude.
- Detect the reference PBD signal as a function of temperature in the temperature range from 16-35 °C with the temperature increment of 3 °C.
- Upon each temperature change, check the position of the probe beam on the position sensitive detector and readjust the position to the center of the detector if necessary.
- Check the linearity of the PBD signal as a function of the [(dn/dt)/Cpρ] term according to Equation 2.
- Place the sample solution in the same optical cell as for the reference compound keeping the same orientation of the optical cell as for the reference measurement.
- Detect the sample PBD traces in the same temperature range as for the reference and check the linearity of the sample PBD amplitude with respect to the [(dn/dt)/Cpρ] term.
4. Data Analysis
The magnitude of the deflection is directly proportional to the volume change due to the sample heating (ΔVth) and nonthermal volume change (ΔVnonth) according to Equation 1: 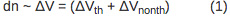
The amplitude of the sample (AS) and reference (Ar) PBD signal can be described using Equations 2 and 3, respectively.
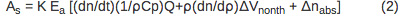
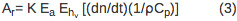
PBD signal is directly proportional to the instrument response parameter (K) and the number of Einsteins absorbed (Ea). The first term in Equation 2, (dn/dt)(1/ρCp)Q, corresponds to the signal change due to the heat released to the solvent. The dn/dt term represents the temperature-dependent change in the index of refraction, ρ is the density of the solvent, Cp is the heat capacity. All parameters are known for the distilled water and can be determined for a buffer solution by comparing a PBD signal for the reference compound in distilled water and in an appropriate buffer. Q is the amount of heat returned to the solvent. The ρ (dn/d ρ ) term is a unit-less constant that is temperature-independent in the temperature range from 10-40 °C 10. Δnabs term corresponds to the change in the index of refraction due to the presence of absorbing species in the solution and it’s negligible if the wavelength of the probe beam is shifted relative to the absorption spectra of any species in the solution. The signal arising from the reference compound (Ar) is expressed by Equation 3 where Ehν is the photon energy at the excitation wavelength, Ehν = 80.5 kcal/mol for 355 nm excitation.
- Take the amplitude of the reference PBD signal as the difference between the pretrigger and post trigger PBD signal as demonstrated in Figure 3. In a similar way, determine the amplitude of the fast (Asfast) and slow phase (Asslow) of the sample PBD signal.
- To eliminate the instrument response parameter, K, scale the amplitude of the sample PBD signal by the amplitude of the PBD signal for the reference. The ratio of the sample signal to the reference signal gives Equation 4 and can be written as:

- Using this equation, determine the heat released to the solution (Q) and nonthermal volume change (ΔVnonth) associated with a photo-initiated reaction from the slope and intercept, respectively, of a plot of [(As/Ar) Ehν] term versus temperature dependent term [(dn/dt)(1/ρCp)].
- To determine the reaction volume and enthalpy change for the fast and the slow process, scale the observed volume and enthalpy change to the appropriate quantum yield according to Equations 5-7.
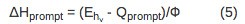
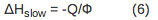
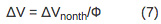
For a multistep process with kinetics occurring on the time-scale between 10 µsec~200 msec, the volume and enthalpy changes associated with the individual steps of the reaction can be determined. The amplitudes and lifetimes for the individual steps are analyzed by fitting the data to the function F(t) that describes the time profile of the volume and enthalpy changes.
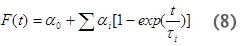
where α0 corresponds to the Asfast and αi corresponds to Asslow for each individual process and τ i are the lifetime of the individual reaction steps. From the temperature-dependence of the rate constant for individual processes (ki = 1/τi) the activation enthalpy and entropy parameters can be readily determined using Eyring plots.
Representative Results
A representative example of PBD traces for Ca2+ photo-release from Ca2+DM-nitrophen is shown in Figure 3. The fast phase corresponds to the photo-cleavage of Ca2+DM-nitrophen and Ca2+ liberation, whereas the slow phase reflects Ca2+ binding to the nonphotolysed cage. The plot of the sample PBD amplitude for the fast and slow phase scaled to the amplitude of the reference compound as a function of the temperature depended factor [Cpρ/(dn/dt)] factor according to Equation 4 and scaling the observed amount of heat released to the solution and nonthermal volume change to the quantum yield for DM-nitrophen photo-dissociation (Φ = 0.18)11 according to Equations 5-7 provided the values of reaction enthalpy and volume for the fast phase (ΔVfast = -12±5 ml/mol and ΔHfast = -50±20 kcal/mol) and for the slow phase (ΔVslow = 9±5 ml/mol and ΔHslow = -23±12 kcal/mol). The PBD trace for the Ca2+ association to the C-terminal domain of the neuronal calcium sensor Downstream Regulatory Element Antagonist Regulator (DREAM) (amino acid residues 161-256) are shown in Figure 4. Upon Ca2+ photo-dissociation, photo-released ligand associates to the C-terminal domain of DREAM with the time constant of 1.3±0.3 msec at 20 °C. From the temperature dependence of the observed rate constant, the activation barrier for Ca2+ binding to the C-terminal domain of DREAM was determined to be 9.2±0.4 kcal/mol.
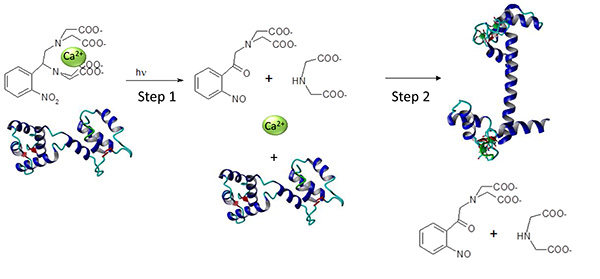
Figure 1. Reaction scheme for the Ca2+DM-nitrohen uncaging and Ca2+ binding to a Ca2+ sensor. Schematic presentation of a photo-initiation of the Ca2+ triggered conformational transition in Ca2+ transducers. Photo-cleavage of Ca2+DM-nitrophen leads to an increase in the free Ca2+ concentration (Protocol 1), Ca2+ association to the Ca2+ transducer and concomitant structural transition (Protocol 2). Click here to view larger image.
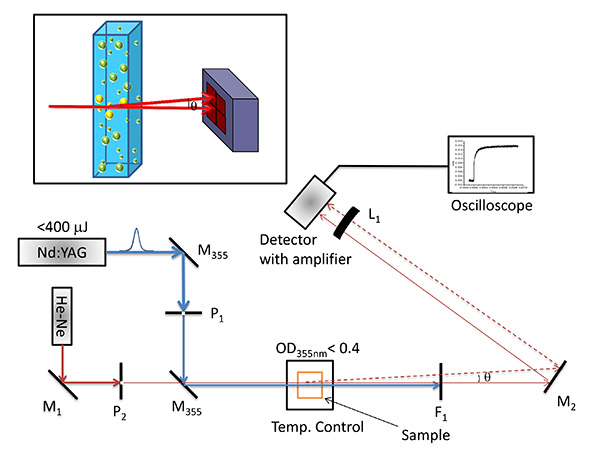
Figure 2. Photothermal beam deflection set-up. Experimental set-up for the photo-thermal beam deflection measurements in the collinear configuration. M1 and M2 represent flat mirrors used to focus the probe beam on the center of a position sensitive detector, M355 represent high energy Nd:YAG laser mirrors, L1 represents a convex lens positioned in front of a detector, P1 and P2 represents pinholes to shape the pump and probe beam diameter, respectively, and DM is a dichroic mirror. F1 represents a 500 nm long pass filter. Inset: Beam deflection due to the photothermal effect. The probe beam that travels through a refractive index gradient created due to a temperature gradient in a medium is deflected by an angle θ and the deflection angle is measured using a position sensitive detector. Click here to view larger image.
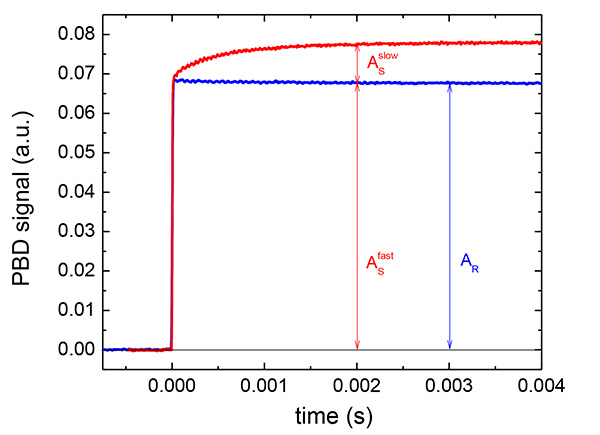
Figure 3. Representative PBD traces for Ca2+DM-nitrophen uncaging. PBD trace for the Ca2+ photo-release release from Ca2+DM-nitrophen (shown in red) and the reference compound (shown in blue). The conditions: 1 mM DM-nitrophen in 20 mM HEPES buffer, 100 mM KCl pH 7.0 and 0.8 mM Ca2+. K3[Fe(CN)6] was used as a reference compound. The absorption of the sample at 355 nm matched that of the reference compound (A355nm = 0.4).The optical density of 0.4 is selected since it is in the range of the linearity of the PBD signal as a function of the number of Einsteins absorbed. The PBD traces represent an average of 20 traces. Click here to view larger image.
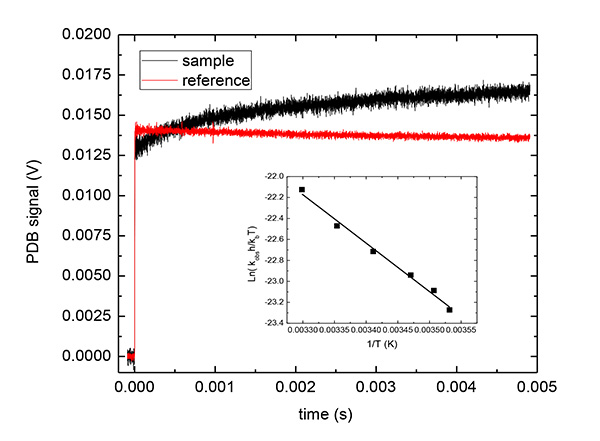
Figure 4. Representative PBD traces for Ca2+DM-nitrophen uncaging in the presence of C-terminal domain of DREAM. PBD trace for Ca2+ photo-release from Ca2+DM-nitrophen in the presence of C-terminal domain of DREAM protein (shown in red) and for the reference compound (shown in black). Inset: Eyring plot using the kinetics recovered from an exponential decay to the sample PBD trace at different temperatures. Conditions 400 μM DM-nitrophen, 390 µM CaCl2, 100 μM C-terminal DREAM in 20 mM HEPES buffer pH 7.4, 100 mM KCl and 500 µM TCEP and trace for the reference under the same conditions. The PBD trace for the reference represent an average of 20 traces and that for the sample is an average of three traces. Click here to view larger image.
Discussion
The physical principle behind photothermal methods is that a photo-excited molecule dissipates excess energy via vibrational relaxation to the ground state, resulting in thermal heating of the surrounding solvent1,12. For solvents such as water, this produces a rapid volume expansion (ΔVth). Excited-state molecules may also undergo photochemical processes that result in nonthermal volume changes (ΔVnonth) due to bond cleavage/formation and/or changes in molecular structure that can alter the molecular dimensions of the molecule(s) (i.e. changes in van der Waals volume), as well as alter the charge distribution of the molecule(s) resulting in electrostriction effects. This results in a rapid expansion of the illuminated volume that generates refractive index change that can be probed by a variety of optical methods. PBD takes advantage of the fact that a refractive index gradient established in the sample due to the Gaussian shaped laser pulse, causes deflection of a probe beam propagated through the sample as demonstrated in Figure 2.
The photo-thermal beam deflection technique enables the determination of time-resolved volume and enthalpy changes for a wide range of photo-initiated processes including the reaction mechanism of photo-cleavage of ligand-heme complexes13 and release of a bioactive molecule from the cage compound complex14 as well as probing the fluorophore triplet state lifetime. This approach offers several advantages compared to traditional techniques commonly used to monitor structural changes in biomacromolecules, such as transient absorption spectroscopy and fluorescence. No intrinsic chromophore/fluorophore or additional protein labeling are required since structural transitions are probed by monitoring changes in volume and/or in enthalpy, although the presence of a chromophore in the sample solution is necessary to generate PBD signal. In addition, kinetic parameters (rate constants, activation enthalpy and entropy) for processes occurring on the submicrosecond timescale, which are not resolved in stop-flow experiments, are easily accessible in PBD measurements. Recently, other label free detection systems that probe a change in the index of refraction or a wavelength shift such as surface plasmon resonance and biolayer interferometry, respectively, were developed and applied to study ligand/protein and protein/protein interactions. However, unlike PBD, these approaches involve immobilization of biological target molecules onto the surface and thus required additional bio-macromolecule modifications of that may interfere with the observed process.
We have adopted several modifications our instrumentation set-up that resulted in better data reproducibility and smaller errors in recovered thermodynamic parameters. Specifically, the application of a quadruple photo-diode as the PBD detector allowed a precise alignment of the probe beam on the position detector and thus better signal reproducibility whereas an additional vibrational isolation of both the probe beam and PBD detector using damped mounting posts decreased the noise of the PBD signal and expanded the time-scales accessible in PBD measurements. We would like to underline that the sensitivity of the experiment depends on the timescale of the event(s)and monitoring of slower processes (τ > 10 msec) is highly sensitive to the instrumentation setup and experimental conditions. Also, placing a dichroic mirror in front of the sample cell simplifies a collinear alignment of the probe and pump beam, increasing the S/N ratio of the PBD signal.
The critical steps of the protocol include regular verification of the linearity of the PBD signal as a function of i) the temperature dependent factor, [(dn/dt)/Cpρ], ii) laser power, and iii) sample/reference absorbance at the excitation wavelength. In addition, precise control of the temperature and identical experimental conditions for sample and reference measurements is crucial for robust PBD measurements and successful data acquisition.
The main limitation of PBD technique reminds the requirement that a studied bio-macromolecule carries a photo-label group. This drawback can be overcome by application of various caged compounds. Indeed, recent development of new caging strategies including caged peptides, caged protein and caged DNA allows for broad application of PBD to monitor complex biological systems and processes including protein – peptide, and protein – protein interactions as well as fast kinetics associated with DNA folding on physiologically relevant time-scales. The strategy presented here clearly demonstrates that in combination with an appropriate cage compound system, the PBD techniques can be easily employed to characterize microsecond to millisecond structural transitions associated with the Ca2+ binding to calcium sensors as well as to monitor small ligands interactions with protein transducers or binding of substrate molecules to enzymes etc.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by National Science Foundation (MCB 1021831, JM) and J. & E. Biomedical Research Program (Florida Department of Health, JM).
Materials
| 1-(4,5-Dimethoxy-2-Nitrophenyl)-1,2-Diaminoethane-N,N,N',N'-Tetraacetic Acid | Life Technologies | D-6814 | DM-nitrophen, cage calcium compound, keep stock solutions in dark to prevent photodissociation, |
| 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid, N-(2-Hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) | Sigma Adrich | 0909C | HEPES buffer |
| Potassium ferricyanide(III) | Sigma Aldrich | 702587 | reference compound for PBD measurments |
| Sodium chromate | Sigma Aldrich | 307831 | reference compound for PBD measurments |
| He-Ne Laser Diode 5mW 635nm | Edmund Optics | 54-179 | use as a probe beam for PBD measurments |
| Oscilloscope, | LeCroy | Wave Surfer 42Xs | 400 MHz bandwith |
| Nd:YAG laser | Continuum | ML II | pump beam for PBD measurments |
| M355; Nd:YAG laser mirror | Edmund Optics | 47-324 | laser mirror for 355 nm laser line |
| M1 and M2; Laser diode mirror | Edmund Optics | 43-532 | visilbe laser flat mirror, wavelength range 300-700 nm |
| P1 and P2; Iris Diaphragm | Edmind Optics | 62-649 | pin hole to shape the probe and pum beams |
| L1; bi-convex lens | Thorlabs | LB1844 | a lens to focus the probe beam at the detector, EFL 50 mm, wavelength range 350 – 2000 nm |
| DM, dichroic mirror | Thorlab | DMLP505 | a longpass dichroic mirror with a cutoff wavelength of 505 nm |
| F1; Edge filter | Andower | 500FH90-25 | a long pass filter with a cutoff wavelength of 500 nm |
| Temperature-controlled cuvette holder | Quantum Northwest | FLASH 300 |
References
- Gensch, T., Viappiani, C. Time-resolved photothermal methods: accessing time-resolved thermodynamics of photoinduced processes in chemistry and biology. Photochem. Photobiol. Sci. 2, 699-721 (2003).
- Larsen, R. W., Mikšovská, J. Time resolved thermodynamics of ligand binding to heme proteins. Coord. Chem. Rev. 251 (9-10), 1101-1127 (2007).
- Westrick, J. A., Peters, K. S. A photoacoustic calorimetric study of horse myoglobin. Bioph. Chem. 37 (1-3), 73-79 (1990).
- Belogortseva, N., Rubio, M., Terrell, W., Miksovska, J. The contribution of heme propionate groups to the conformational dynamics associated with CO photodissociation from horse heart myoglobin. J. Inorg. Biochem. 101 (7), 977-986 (2007).
- Mikšovská, J., Suquet, C., Satterlee, J. D., Larsen, R. W. Characterization of Conformational Changes Coupled to Ligand Photodissociation from the Heme Binding Domain of FixL. Biochimie. 44 (30), 10028-10036 (2005).
- Miksovska, J., Gennis, R. B., Larsen, R. W. Photothermal studies of CO photodissociation from mixed valence Escherichia coli cytochrome bo3. FEBS Lett. 579 (14), 3014-3018 (2005).
- Losi, A., Michler, I., Gärtner, W., Braslavsky, S. E. Time-resolved Thermodynamic Changes Photoinduced in 5,12-trans-locked Bacteriorhodopsin. Evidence that Retinal Isomerization is Required for Protein Activation. Photochem. Photobiol. 72, 590-597 (2000).
- Kondoh, M., et al. Light-Induced Conformational Changes in Full-Length Arabidopsis thaliana Cryptochrome. J. Mol. Biol. 413 (1), 128-137 (2011).
- Kaplan, J. H., Ellis-Davies, G. C. Photolabile chelators for the rapid photorelease of divalent cations. Proc. Natl. Acad. Sci. U.S.A. 85 (17), 6571-6575 (1988).
- Eisenberg, H. Equation for the Refractive Index of Water. J. Chem. Phys. 43 (11), 3887-3892 (1965).
- Ellis-Davies, G. C., Kaplan, J. H., Barsotti, R. J. Laser photolysis of caged calcium: rates of calcium release by nitrophenyl-EGTA and DM-nitrophen. Biophys. J. 70, 1006-1016 (1996).
- Miksovska, J., Larsen, R. W. Structure-function relationships in metalloproteins. Methods Enzymol. 360, 302-329 (2003).
- Miksovska, J., Norstrom, J., Larsen, R. W. Thermodynamic profiles for CO photodissociation from heme model compounds: effect of proximal ligands. Inorg. Chem. 44 (4), 1006-1014 (2005).
- Dhulipala, G., Rubio, M., Michael, K., Miksovska, J. Thermodynamic profile for urea photo-release from a N-(2-nitrobenzyl) caged urea compound. Photochem. Photobiol. Sci. 8, 1157-1163 (2009).

